Task: To build a lightweight multimodal medical image fusion network and implement a novel Structural Similarity Pseudo labels Iteration (SPI) mechanism for network training, which continuously refines pseudo labels through iterative screening, thus considering these labels as ground truth in supervised training mode, and therefore addressing the heterogeneity between fused images and source images, which is typically observed in most existing image fusion networks relying on unsepervised training, and making the entire network optimization more effective.
Rationale: The field of multimodal medical image fusion currently lacks ground truth labels (perfect fused images), leading most existing fusion methods to rely on unsupervised training approaches that require substantial prior knowledge and complex loss function design. To address these challenges, we propose a novel Structural Similarity Pseudo labels Iteration (SPI) mechanism for network training, which continuously refines pseudo labels through iterative screening. This approach utilizes both initial labels and progressively augmented pseudo labels data to constrain network optimization, overcoming the limitations of fixed loss functions.
Description: The LWNet and proposed SPI mechanism employs the Structural Similarity Pseudo Labels Iterations (SPI) mechanism for network training using pseudo labels as ground truth to address the heterogeneity between fused images and source images, making the entire network optimization more effective. In addition LWNet adopts a real-time network structure employing adaptive channel attention (ACA) and a multi-scale cross attention (MCA) mechanism in order not only to enhance the retention of important information across channels but also capture stronger global feature dependencies compared to traditional attention mechanisms. The ACA mechanism prioritizes information-rich channels for feature fusion, and the MCA module decomposes and reassembles complementary heterogeneous input images to capture long-range feature dependencies.
Evaluation: All datasets for evaluation were obtained from the Harvard Medical School public dataset (Whole Brain Atlas), which includes brain images of normal brains and various brain diseases. The developers selected 320 pairs of MRI-SPECT images and cropped them into 15,680 120 × 120-pixel patches as the training set, where 8000 patches were chosen as labeled data, and the remaining 7680 patches were iteratively selected as unlabeled labels. Additionally, 37 pairs of MRI-SPECT and 30 pairs of MRI-PET images were chosen as the test set. Since the format of the MRI-PET dataset is similar to that of MRI-SPECT, the developers directly generalized the pre-trained model to the MRI-PET test set. For the MRI-CT experiment, 154 pairs of MRI-CT images were selected and similarly cropped into 7546 120 × 120-pixel patches as the training set, where 4000 patches were chosen as labeled data, and the remaining 3546 patches were iteratively selected as unlabeled labels. Finally, 30 pairs of MRI-CT images were chosen as the test set.
The training experiments were conducted on a server with the following configuration: a 2.30 GHz Intel(R) Xeon(R) Gold 5218 CPU, 187 GB RAM, and two Nvidia GTX2080ti GPUs. The proposed method was implemented using the PyTorch 1.12.1 framework, with the Adam optimizer used for training. The hyperparameters and were set to 1.10 and 1.00, respectively, and the batch size was set to 4. The training process consisted of 10 iterations, with each iteration involving 10 epochs of supervised training. Testing was conducted on a personal computer with the following configuration: a 3.00 GHz Intel(R) Core(TM) i5-12490F, 32 GB RAM, and an Nvidia GTX4060ti GPU.
To validate the effectiveness of the proposed method, the developers selected nine comparison methods, including two traditional methods and seven deep learning methods (where LRD and EgeFusion are traditional methods), as shown in the Table below:

Abbreviations of comparison methods and their corresponding published journals.
To objectively demonstrate the advantages of the proposed method, we evaluated it from multiple perspectives, such as information content, gradient, and signal-to-noise ratio. Ultimately, we selected the following six objective metrics: Variance Inflation Factor (VIF) [Ext. Ref.], Mutual Information (MI) [Ext. Ref.], Universal Quality Index (UQI) [Ext. Ref.], Structural Similarity Index Metric (SSIM) [Ext. Ref.], Peak Signal-to-Noise Ratio (PSNR) [Ext. Ref.], and Chen-Blum metric (Qcb) [Ext. Ref.]. All metrics indicate that higher values correspond to higher quality of the fused images.
Results: The LWNet frameworkj with the SPI mechanism is quantitatively and qualitatively analyzed on multiple datasets demosntrating significant advantages over state-of-the-art methods. Furthermore, LWNet exhibits the lowest structural complexity meeting the requirements for low latency and high responsiveness in medical applications.
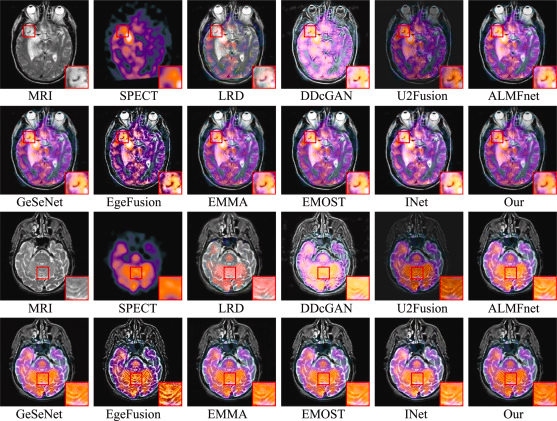
Regarding MRI-SPECT fusion and the developers’ quantitative analysis, they selected two pairs of MRI-SPECT fused images for visualization, as shown in the Figure below. The first and second rows correspond to the first image pair, while the third and fourth rows represent the second image pair. Firstly, the LRD fusion method exhibits severe false color distortion. The DDcGAN method struggles to balance the loss between one generator and two discriminators, resulting in a significant loss of internal detail information. In the case of U2Fusion, the fused images clearly suffer from insufficient brightness, which may increase the difficulty of downstream tasks. As EdgeFusion is based on edge information, its results show overly sharp edges and introduce a considerable amount of noise during the fusion process. Although some other methods perform well in preserving details, they often suffer from overexposure or underexposure in brightness compared to our fusion approach. Therefore, from a visual perspective, LWNet (labeled as “Our” in the figure) achieves superior performance in both detail preservation and brightness balance.
Visualization of two pairs of fused images from the MRI-SPECT dataset for LWNet and comparison methods.
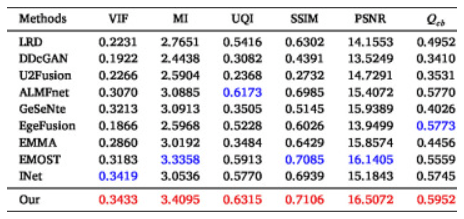
For quantitative analysis of the MRI-SPECT fusion results, as shown in the table below, the proposed LWNet method ranks second in the MI metric but falls behind the state-of-the-art method by only 0.01. For the remaining five metrics, LWNet achieves the best results, significantly outperforming state-of-the-art methods, particularly in UQI, SSIM, and Qcb. Overall, LWNet outperforms the state-of-the-art by 0.7%, 2.6%, 1.9%, 4.7%, and 4.1% in terms of VIF, UQI, SSIM, PSNR, and Qcb, respectively.
The average values of six metrics on the MRI-SPECT dataset. Red:The Best, Blue:The Second Best
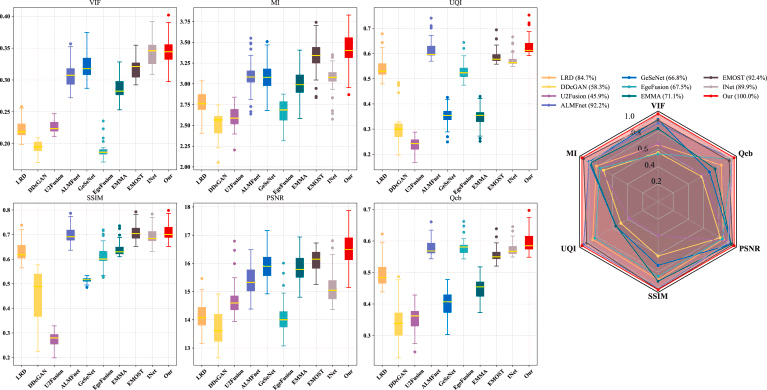
As shown in the figure below, box plots were employed to analyze the distribution of metrics across all images in the testset. Evidently, the majority of our data points are positioned at the top among all methods, indicating superior fusion performance for each individual image. Additionally, radar charts (with all metrics normalized) are used to evaluate the comprehensive capability of the models. The percentage values represent the area ratio of the hexagon enclosed by the metrics relative to the total hexagon area — larger values denote stronger fusion performance. Visually, our proposed method demonstrates optimal performance in both area ratio and proximity of data points to the vertices.
Boxplot and radar chart visualization of data distribution for each image in the MRI-SPECT dataset
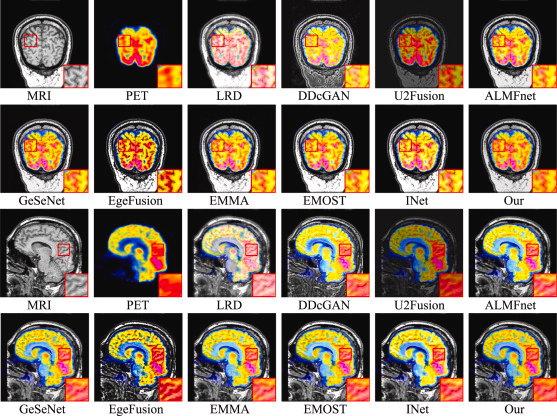
The equivalent results for the MRI-PET fusion data are shown below:
Visualization of two pairs of fused images from the MRI-PET dataset for LWNet and comparison methods
The average values of six metrics on the MRI-PET dataset. Red:The Best, Blue:The Second Best.
Boxplot and radar chart visualization of data distribution for each image in the MRI-PET dataset.
Claim: A novel Structural Similarity Pseudo labels Iteration (SPI) mechanism for supervised medical image fusion network training is proposed. SPI utilizes fusion results from multiple state-of-the-art methods as ground truth labels. During the iterative process, appropriate fused images are selected as pseudo labels based on structural similarity metrics with predefined thresholds. The combined supervision from both ground truth and pseudo labels overcomes the limitation of conventional methods that only constrain the relationship between fused and source images.
Furthermore, a lightweight multimodal medical image fusion network is introduced incorporating (i) an Adaptive Channel Attention (ACA) mechanism to enhance feature extraction by suppressing attention to less important channels, and (ii) a Multi-scale Cross-Attention (MCA) module that establishes global feature dependencies through cross-attention computation of features cropped by different convolutional kernels, thereby reducing noise introduction during fusion. Experimental results demonstrate that the LWNet fusion model achieves superior visual quality and objective evaluation metrics compared to existing medical image fusion approaches.
Remarks: The performance evaluation study of the LWNet developers may have certain limitations. Since both the acquisition of ground truth labels and the selection of pseudo labels are based on multimodal medical images, LWNet may not be directly applicable to other fusion tasks such as infrared and visible image fusion or multi-focus image fusion. In such cases, new ground truth labels would need to be selected and the model retrained from scratch. Furthermore, the current ground truth labels used in the evaluation study are derived from several state-of-the-art fusion methods in recent years. However, this approach is inherently limited, as these methods may themselves contain imperfections, which could be inadvertently learned by the model during training.
In future work, the LWNet developers plan to design more sophisticated iterative mechanisms to enhance the effectiveness of network training constraints. When selecting high-quality pseudo labels, the aim is to go beyond structural similarity metrics by incorporating manually annotated textual datasets to assist in the selection process. Additionally, for ground truth selection, manually curated high-quality fused images can be used as ground truth to improve the effectiveness of the initial training phase. Regarding network architecture, more advanced or lightweight modules can be integrated into the overall structure to achieve better fusion performance with reduced computational time.
Data Availability Statement: Data will be made available by the developers on request.