Task: Multimodal (PET & multi-modal MR) 3D medical image synthesis
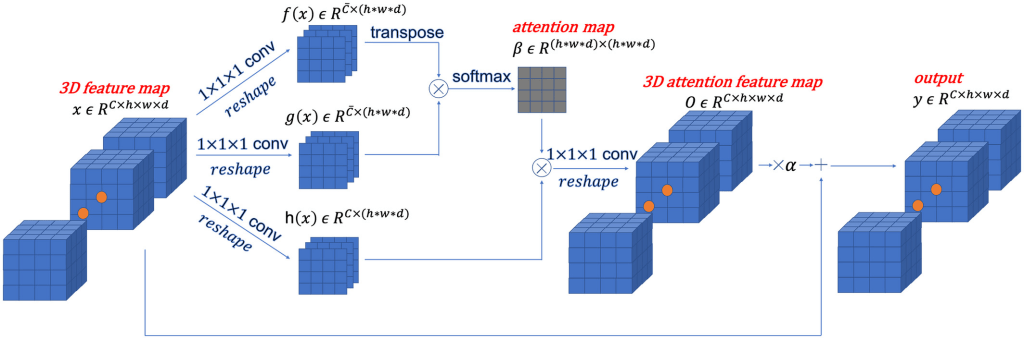
Description: A novel 3D conditional generative adversarial network (GAN) designed and optimized for the application of multimodal 3D neuroimaging synthesis. The model uses spectral normalization and feature matching to stabilize the training process and ensure optimization convergence (SC-GAN). A self-attention module was also added to model the relationships between widely separated image voxels.
The model is constructed as follows: First, a 2D conditional GAN is extended into a 3D conditional GAN (cGAN). Next, a 3D self-attention module is added to generate 3D images with preserved brain structure and reduced blurriness in the synthesized images. This module models the similarity between adjacent and widely separated voxels of a 3D image. Spectral normalization, feature matching loss and brain area RMS error (RMSE) loss are also introduced to stabilize the network training process and prevent overfitting.
The novel features of the SC-GAN model is:
- introduction of a 3D self-attention module into 3D conditional GAN to generate high-accuracy synthesis results with stable training process. A smooth training is then achieved by using of a series of stabilization techniques and a modified loss function; and
- testing on multiple data sets across different synthesis tasks and enabling of multimodal input, which can be generalized to a wide range of image synthesis applications.
Validation: The performance of the network was evaluated on the ADNI-3 dataset, in which the proposed network was used to predict amyloid beta PET (Aβ-PET) images, fractional anisotropy (FA), and mean diffusivity (MD) maps from multimodal MRI. Then, SC-GAN was applied on a multidimensional diffusion MRI experiment for super-resolution application. Experiment results were evaluated by normalized RMS error, peak SNR, and structural similarity. Three image-quality metrics were used to evaluate the performance of the synthesis task: normalized RMSE (NRMSE), peak SNR, and structural similarity.
Results: In general, SC-GAN outperformed other state-of-the-art GAN networks including 3D conditional GAN in all three tasks across all evaluation metrics. Prediction error of the SC-GAN was 18%, 24% and 29% lower compared to 2D conditional GAN for fractional anisotropy, PET and mean diffusivity tasks, respectively. The ablation experiment showed that the major contributors to the improved performance of SC-GAN are the adversarial learning and the self-attention module, followed by the spectral normalization module. In the super-resolution multidimensional diffusion experiment, SC-GAN provided superior predication in comparison to 3D Unet and 3D conditional GAN.
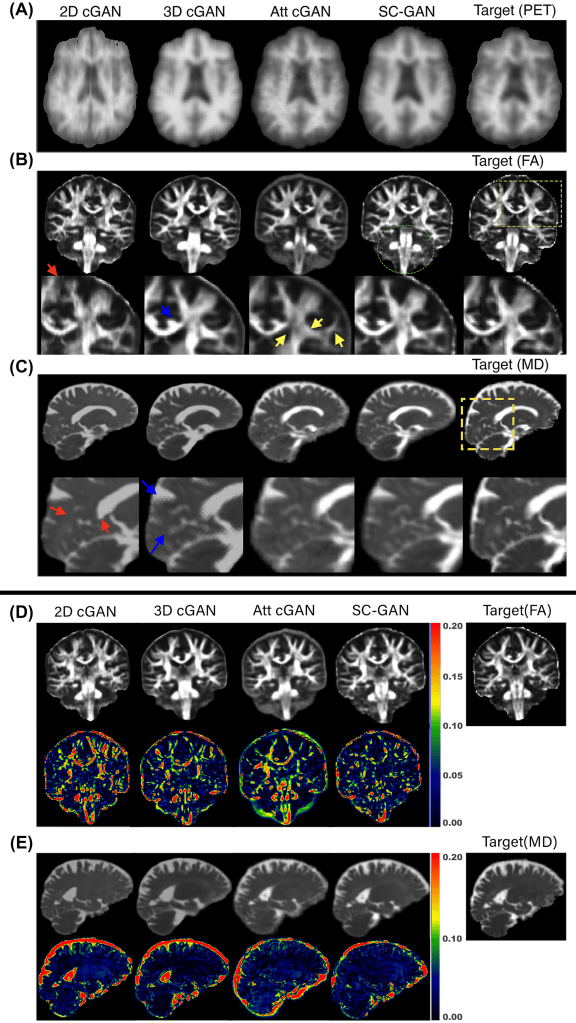
Claim: The SC-GAN network is an end-to-end medical image synthesis network that can be applied to high-resolution, multimodal input images. The SC-GAN was designed to handle multimodal (multichannel) 3D images as inputs. It was shown that SC-GAN significantly outperformed state-of-the-art techniques, enabling reliable and robust deep learning–based medical image synthesis for a wide range of applications.
The high computational cost is the main limitation of SC-GAN; it requires lengthy training and heavy computational resources, such as GPU memory. Sparse attention matrix computation could be a potential solution. A future research direction could focus on generalizing SC-GAN, such as by adopting knowledge distillation mechanism into SC-GAN.
Data Availability: The Alzheimer’s Disease Neuroimaging Initiative ADNI-3 dataset was used. The tools and codes used in this project are mostly available through the Laboratory of Neuroimaging (LONI) pipeline. The FreeSurfer and FSL open toolkits were employed for preprocessing and data preparation. The proposed SC-GAN deep learning network is also made publicly available.
