Task: Build a model (BAP2A-public) to predict brain age using 5-HT2AR binding PET outcomes, compare 5-HT2AR-PET-based predictions of brain age to predictions based on gray matter (GM) volume, as determined with structural magnetic resonance imaging (MRI), and investigate whether combining 5-HT2AR and GM volume data improves prediction
Rationale: To better assess the pathology of neurodegenerative disorders and the efficacy of neuroprotective interventions, it is necessary to develop biomarkers that can accurately capture age-related biological changes in the human brain. Brain serotonin 2A receptors (5-HT2AR) show a particularly profound age-related decline and are also reduced in neurodegenerative disorders, such as Alzheimer’s disease.
Machine learning (ML) allows for modeling the healthy aging brain by summarizing aging brain biology from neuroimages into one single variable, called “brain age” [Ext. Ref., Ext. ref.]. Models are trained to predict chronological age on imaging data from healthy individuals based on structural or functional changes that occur over the human lifespan. The result is a model of a healthy aging brain, indicating how an average healthy brain would change during an individual’s lifespan. More than the predicted age itself, the deviation of the predicted age from the true chronological age is of interest. This deviation is hypothesized to reflect divergence from the expected healthy aging trajectory [Ext. Ref.]. The deviation between predicted age (A_pred) and chronological age (A_chrono) is expressed as the predicted age deviation (PAD)
PAD = A_pred – A_chrono
Under this hypothesis, a higher prediction than chronological age reflects a biologically older brain. Following from this, a positive PAD should translate to an increased risk for aging-related disorders. An increased PAD has been linked to increased cardiovascular risk [Ext. Ref.], overall mortality [Ext. Ref.], neurodegenerative disorders, and cognitive decline [Ext. Ref., Ext. Ref., Ext. Ref., Ext. Ref., Ext. Ref.].
The model developers aimed to investigate if the age-related decline in 5-HT2AR binding can be utilized in the brain age paradigm. Specifically, the first aim is to apply ML algorithms to 5-HT2AR binding outcomes from PET images to predict brain age. The second aim is to compare these estimates to those derived from applying the same algorithms to volumetric GM data derived from structural MR images. The third aim is to investigate whether a multimodal approach combining 5-HT2AR binding and GM measures improves the prediction of brain age.
Description: To develop a predicitve Machine Learning (ML) model of the healthy aging brain, a set of ML algorithms were trained to predict chronological age from imaging-derived GM volume (MRI) and 5-HT2AR (PET) binding data. The algorithms were implemented using scikit-learn (v1.2) [Ext. Ref.] in python 3.9.12. Commonly used algorithms were selected for brain age prediction [Ext. Ref., Ext. Ref.]: (1) Bayesian Ridge Regression (BRidge), (2) Relevance Vector Regression (RVR) implemented using ARDRegression, (3) Gaussian Process Regression with linear kernel (linGPR), (4) Gaussian Process Regression with radial basis function kernel (rbfGPR), and (5) linear support vector regression (linSVR). The selected algorithms were trained on either 5-HT2AR binding outcomes, GM volumes, or both. The multimodal model, combining structural MRI and 5-HT2AR PET-derived data, was implemented as a stacking regressor [Ext. Ref.]. In a stacking regressor, a base model for each modality was trained, with outcomes then used as input into a linear regression model. The algorithms BRidge, RVR, linGPR, linSVR, and rbfGPR were used as a base estimator, resulting in five ensemble regressors.
Two references were implemented to put the results into perspective. First, a dummy regressor ws trained to always output the mean age of the training. Second, pyment was applied, which is a pre-trained state-of-the-art structural MRI-based brain age prediction software. pyment uses a skull-stripped and MNI-space registered T1w-image as input [Ext. Ref.]. It was originally trained on images from 53,542 healthy individuals and has been shown to predict age on unseen data with high accuracy and reliability [Ext. Ref.].
The chronological age was predicted based on the three feature sets (5-HT2AR, GM, and 5-HT2AR + GM) as well as the two reference models. An overview of the respective prediction pipelines is presented in the figure below.
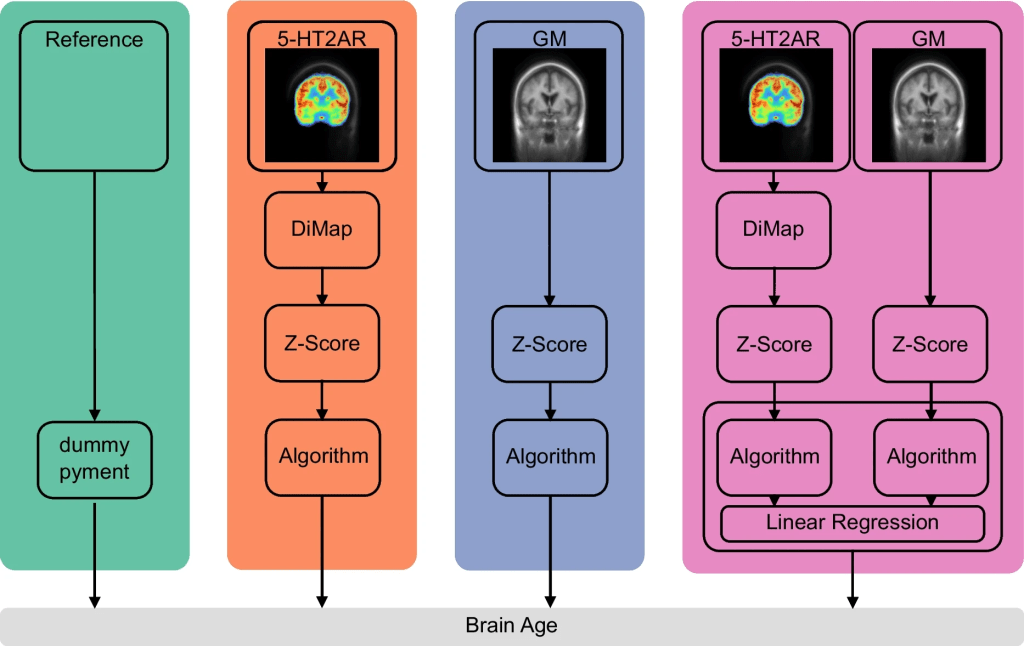
Overview of the prediction pipelines. Reference: the pre-trained MRI-based prediction model pyment and a dummy regressor always predicting the mean chronological age of the training data. 5-HT2AR: ([11C]Cimbi-36) is transformed to -like units using the distribution mapping (DiMap). BPs are then z-scored. GM: volumes of cortical and subcortical regions are z-scored. 5-HT2AR + GM: transformations for 5-HT2AR and GM features are identical to the unimodal counterparts. Brain age is predicted using a stacking ensemble method where a base model is trained for each modality, with the outcome used as input into a final estimator
The different outcome measures for [11C]Cimbi-36 and [18F]altanserin were aligned by transforming regional 5-HT2AR to -like values based on age-matched subsets using a distribution mapping approach [Ext. Ref.]. Subsequently, the matched values were standardized (zero mean, unit variance). GM volumes were similarly z-scored. A multimodal brain age estimate was calculated combining 5-HT2AR binding and GM volume features using the stacking approach previously described. Each feature set was transformed as described before.
Evaluation: The BAP2A-public model is evaluated by using PET and MR images from 209 healthy individuals aged between 18 and 85 years (mean=38, std=18), and estimated 5-HT2AR PET-based binding and GM MR-based volume for 14 cortical and subcortical regions. Different machine learning algorithms were applied to predict chronological age based on 5-HT2AR binding, GM volume, and the combined measures. The mean absolute error (MAE) and a cross-validation approach were used for evaluation and model comparison.
All PET images were corrected for inter-frame motion using the AIR algorithm (v5.3.0) [38]. The MR images have been corrected for spatial distortions due to nonlinearities in gradient fields. This preprocessing was part of the Cimbi database curation.
Standard FreeSurfer (v7.2) segmentation and parcellation pipelines [Ext. Ref.] were applied to the MR images to retrieve masks for cortical and subcortical regions of interest (ROIs) from the Desikan-Killiany [Ext. Ref.] and Aseg atlas [Ext. Ref.]. Cortical ROIs were grouped into seven larger cortical regions (frontal lobe, occipital lobe, parietal lobe, temporal lobe, parahippocampal gyrus, cingulate, and insula) [Ext. Ref.] with high 5-HT2AR binding, as identified by autoradiography [Ext. Ref., Ext. Ref.]. Additionally, seven subcortical brain regions were used in the analysis (thalamus, caudate, putamen, pallidum, hippocampus, amygdala, and nucleus accumbens). All regions were averaged across hemispheres, resulting in 14 ROIs. The MR images were co-registered to a summed PET image using SPM8. The resulting co-registration matrix was used to project the ROI masks on the dynamic PET files to extract time-activity curves (TACs) for each region.
The volume of all ROIs was extracted from the FreeSurfer output and normalized using the intracranial volume [Ext. Ref.]. Regional binding potentials (BPs) were calculated using the regional TACs extracted from the PET data. The cerebellum excluding vermis was used as a reference region, as it has a negligible density of 5-HT2ARs [Ext. Ref.]. 5-HT2AR binding from [11C]Cimbi-36 scans was quantified as the non-displaceable binding potential (BP_nd) using the multilinear reference tissue model 2 [Ext. Ref.], with k2′ values estimated using the simplified reference tissue model applied to a TAC extracted from a mask over the entire neocortex. Quantification of [18F]altanserin binding has been described in detail elsewhere (Pinborg et al., 2003). In short, the ratio of specifically bound radioligand to that of total parent radioligand in plasma, (BP_p) was calculated and used as the outcome measure. Even though BP_p and BP_nd are different measures, both are proportional to the density of the 5-HT2AR and highly correlated [Ext. Ref.].
Model accuracy was assessed using the mean absolute error (MAE), calculated as the average absolute difference between predicted and chronological age for each validation fold. The generalization error of each model was estimated based on the 100 hold-out folds in a 20-times repeated five-fold cross-validation (CV) setup [Ext. Ref.]. An additional inner five-fold CV was used for hyperparameter tuning, if necessary. The initial split was randomly shuffled for each of the 20 repetitions to gain a split-independent estimate of the generalization error. The outer five-fold CV was stratified by age to ensure the same age distribution between the training and validation sets.
To assess whether GM volumes are more predictive of age than 5-HT2AR binding in the same regions and whether a multimodal approach (combining 5-HT2AR + GM features) can improve predictions, the best model trained on GM or 5-HT2AR + GM features was tested against the best model trained on only 5-HT2AR features using a paired two-tailed t-test. Specifically, the MAE per fold for each model was used for comparison. As most of the training data overlaps in each CV split, the assumption of independence between the CV folds within each repetition was violated. Therefore, a correction to the t-test was applied to adjust for this dependence [Ext. Ref., Ext. Ref.]. The significance level was set to 0.05.
To investigate the similarity between predictions, the correlation of predictions of the models based on GM volumes and 5-HT2AR binding was calculated. A high correlation coefficient would indicate that the information in both feature sets leads to similar brain age predictions. As an underlying strong correlation with chronological age is expected, the PAD values from the different feature sets were also correlated. Additionally, the weight for each modality in the stacking process of the combined model was investigated. This indicates which feature set contributes most to the final predictions of the combined model.
The BRidge algorithm allows for easy retrieval and interpretation of fitted model weights. These weights indicate how much each feature contributed, i.e., how important the feature was, to the final prediction. The fitted model weights were obtained and averaged over CV folds and then displayed for each ROI on a standard T1w template image. This analysis was performed post hoc.
Results: Findings suggest that both the cerebral 5-HT2AR binding (mean MAE=6.63 years, std=0.74 years) and GM volume (mean MAE=6.95 years, std=0.83 years) predict chronological age accurately. Combining the two measures improves the prediction further (mean MAE=5.54 years, std=0.68).
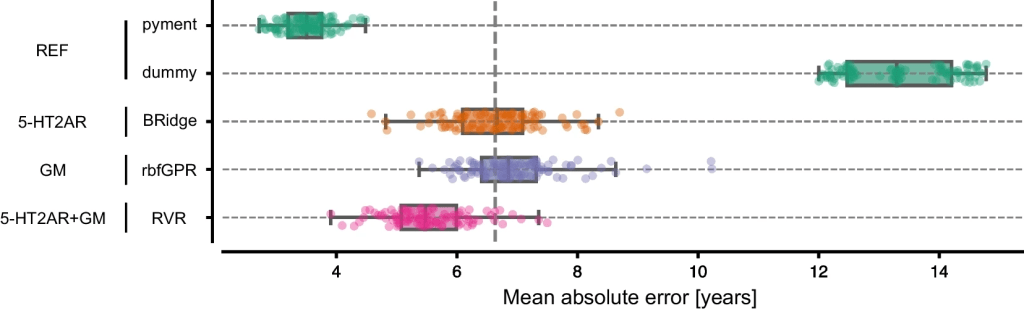
MAE for the best-performing models for each feature set. Each dot represents the MAE for one of the 100 CV iterations. The vertical dashed line marks the generalization error (mean over MAE for each fold) of the 5-HT2AR model
The figure below shows the mean prediction for each subject over the 20 iterations on each feature set for the specific best performing model. There was a strong correlation between chronological and predicted age and a moderate to strong negative correlation between PAD and chronological age.
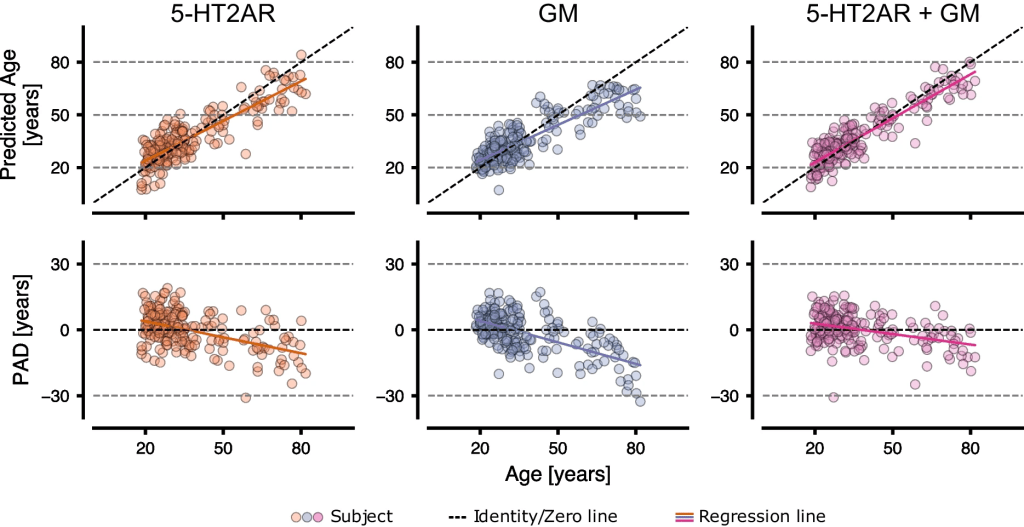
The predicted age and PAD vs. the chronological age for 5-HT2AR, GM, and 5-HT2AR + GM using the best performing model (5HT2AR: BRidge; GM: rbfGPR; 5HT2AR + GM: RVR). Each point represents a mean value over 100 CV iterations. The colored line represents the fitted regression line
The contributions of each modality and ROI to the brain age predictions were also investigated. For the sake of clarity, the results for the Bayesian Ridge Regressor were used in this analysis, since it is a linear model and weights are easily derived. The correlation between the PAD from the models based on 5-HT2AR binding vs. GM volume feature sets was r = 0.35 (R2 = 0.12) as indicated in section A of the below figure. Section B shows the weight of each feature set in the ensemble regressor, with 5-HT2AR-based predictions contributing more to the final outcome. The contribution from individual ROIs to the predicted age for each base model is visualized in section C. For 5-HT2AR binding, the temporal and parietal lobes, insula, caudate, and putamen displayed the highest weights. The frontal lobe and insula contributed the most to age prediction based on GM volumes.
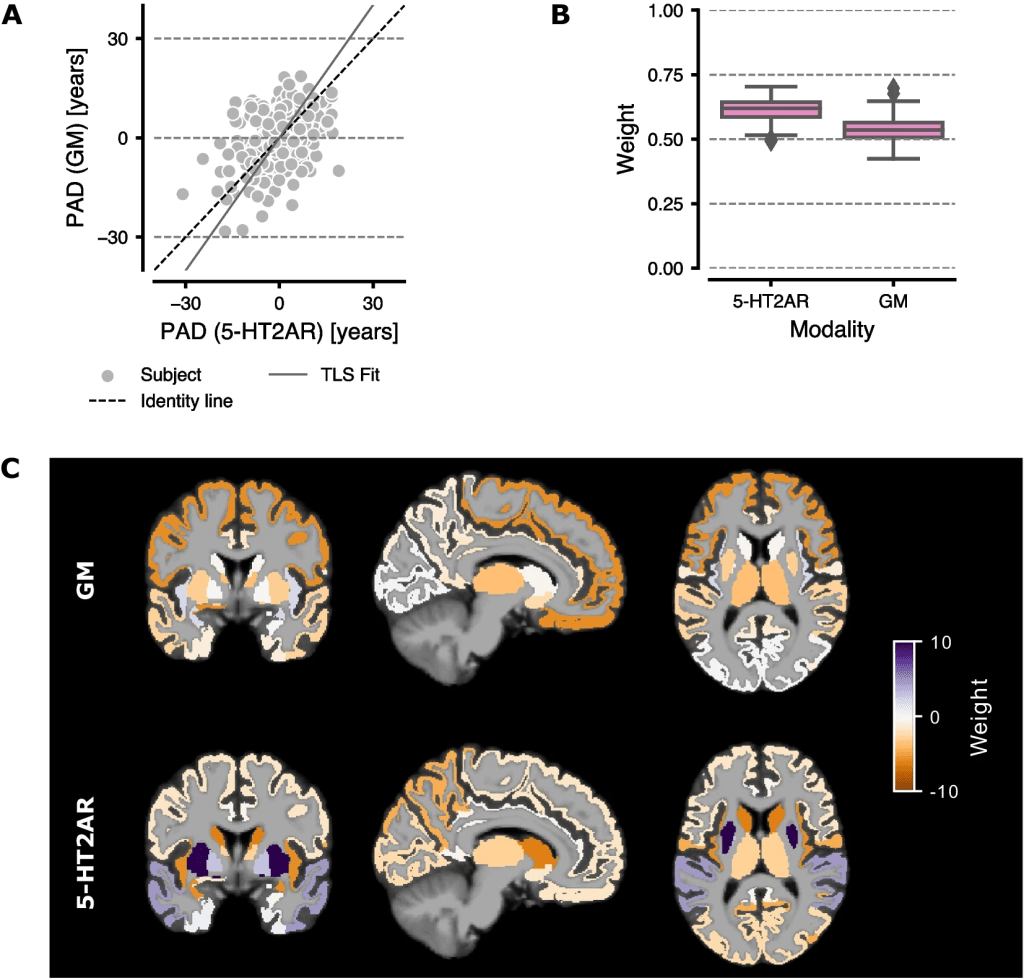
Comparison of 5-HT2AR vs GM-based models. A The PAD from GM volume-based predictions vs 5-HT2AR binding predictions. The solid gray line marks the regression line fitted using total least squares (TLS). Each point represents the mean PAD per subject averaged over all 100 CV iterations. B Boxplots showing the weights for the final ensemble regressor. Whiskers show 1.5 the interquartile range. C The weights for the ensemble base regressors (BRidge) overlaid on a MNI152 template brain
Claim: The BAP2A-public model developers conducted a study to investigate whether 5-HT2AR binding can be used as a putative biomarker for brain aging. For this purpose, they trained ML algorithms to predict chronological age based on regional binding estimates from 5-HT2AR PET images. They also trained ML models to predict chronological age from MRI GM volume estimates from the same regions, and we investigated whether combining both feature sets could improve overall performance. They found that ML algorithms trained on regional 5-HT2AR binding predicted chronological age with similar accuracy to those using regional GM volume as input. Further, combining data from both feature sets significantly improved the overall performance compared to both unimodal models. 5-HT2AR binding measured using PET might be useful for improving the quantification of a biomarker for brain aging.
Remarks: The reported results are based on data from healthy individuals, and conclusions from the current study are hence limited to a healthy population. However, previous PET studies have shown that, compared to age-matched healthy controls, cerebral 5-HT2AR binding is reduced in mild cognitive impairment and in Alzheimer’s disease [Ext. Ref., Ext. Ref.]. This suggests that a brain age model based on 5-HT2AR binding could be useful in predicting neurodegeneration. Future studies should investigate whether 5-HT2AR-based brain age estimation is useful as a biomarker for pathological aging and can be used for e.g., early detection of neurodegenerative disorders or for evaluating putative neuroprotective interventions intended to slow age-related processes in the human brain.
Data Availability: The data that support the findings of this study are available on request from the CIMBI database and an appropriate data-sharing agreement. The data are not publicly available due to privacy or ethical regulatory restrictions.
bioRxiv (preprint)
