Task: TMTV-Net, is specifically designed to automate TMTV segmentation from PET/CT scans. TMTV-Net demonstrates robust performance and adaptability in TMTV segmentation across diverse multi-site external datasets, encompassing various lymphoma subtypes.
Public Availability: TMTV-Net model is containarized in GitHub for additional multi-site evaluations and generalizability analyses. The developed model and the necessary pre-processing steps are made available for multi-site testing and clinical evaluation through a cloud-based platform in GitHub, which is user-friendly and requires no coding.
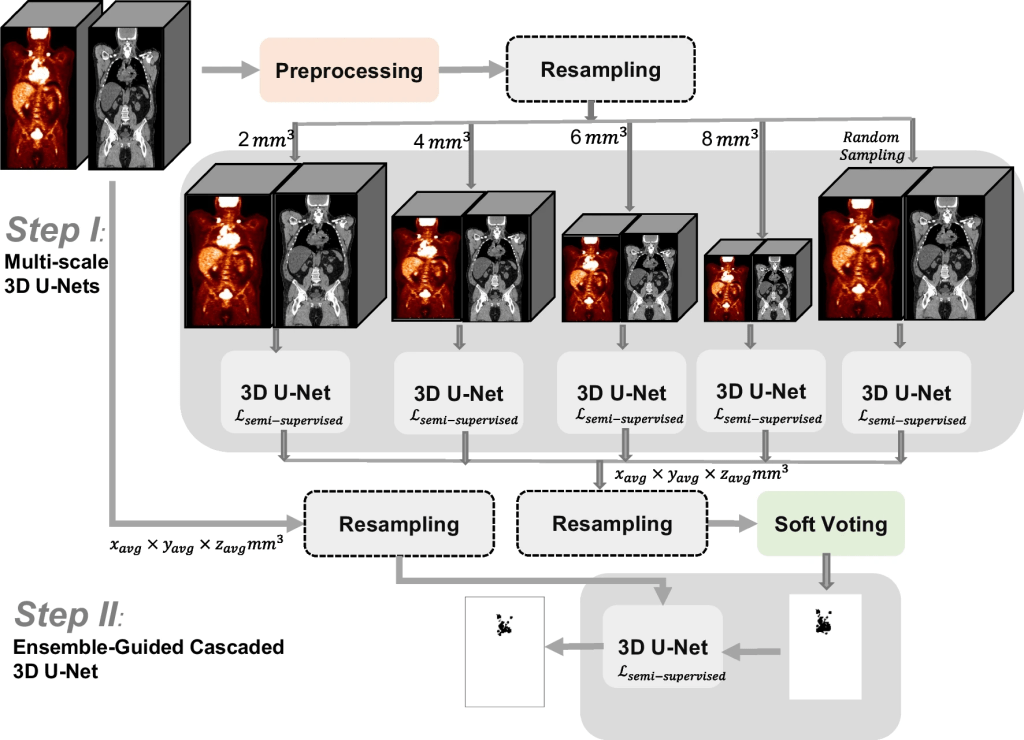
Description: Total metabolic tumor volume (TMTV) segmentation has significant value enabling quantitative imaging biomarkers for lymphoma management. TMTV-Net is designed to adress the challenging task of automated tumor delineation in lymphoma from PET/CT scans using a cascaded approach. This involves resampling PET/CT images into different voxel sizes in the first step, followed by training multi-resolution 3D U-Nets on each resampled dataset using a fivefold cross-validation scheme. The models trained on different data splits were ensemble. After applying soft voting to the predicted masks, in the second step, the probability-averaged predictions were fed, along with the input imaging data, into another 3D U-Net. Models were trained with semi-supervised loss. The effectiveness of using test time augmentation (TTA) was also considred to improve the segmentation performance after training.
Overview of the two-stage cascaded approach for TMTV segmentation, TMTV-Net
Evaluation: Quantitative analysis including Dice score (DSC) and TMTV comparisons was performed. In addition, the qualitative evaluation was also conducted by nuclear medicine physicians.
Results: The cascaded soft-voting guided approach resulted in performance with an average DSC of 0.68 ± 0.12 for the internal test data from developmental dataset, and an average DSC of 0.66 ± 0.18 on the multi-site external data (n = 518), significantly outperforming (p < 0.001) state-of-the-art (SOTA) approaches including nnU-Net and SWIN UNETR. While TTA yielded enhanced performance gains for some of the comparator methods, its impact on our cascaded approach was found to be negligible (DSC: 0.66 ± 0.16). Our approach reliably quantified TMTV, with a correlation of 0.89 with the ground truth (p < 0.001). Furthermore, in terms of visual assessment, concordance between quantitative evaluations and clinician feedback was observed in the majority of cases. The average relative error (ARE) and the absolute error (AE) in TMTV prediction on external multi-centric dataset were ARE = 0.43 ± 0.54 and AE = 157.32 ± 378.12 (mL) for all the external test data (n = 518), and ARE = 0.30 ± 0.22 and AE = 82.05 ± 99.78 (mL) when the 10% outliers (n = 53) were excluded.

Comparison of TMTV-Net 3D segmentation approach to SOTA approaches on a DLBCL case (GT: ground truth, MIP: maximum intensity projections)
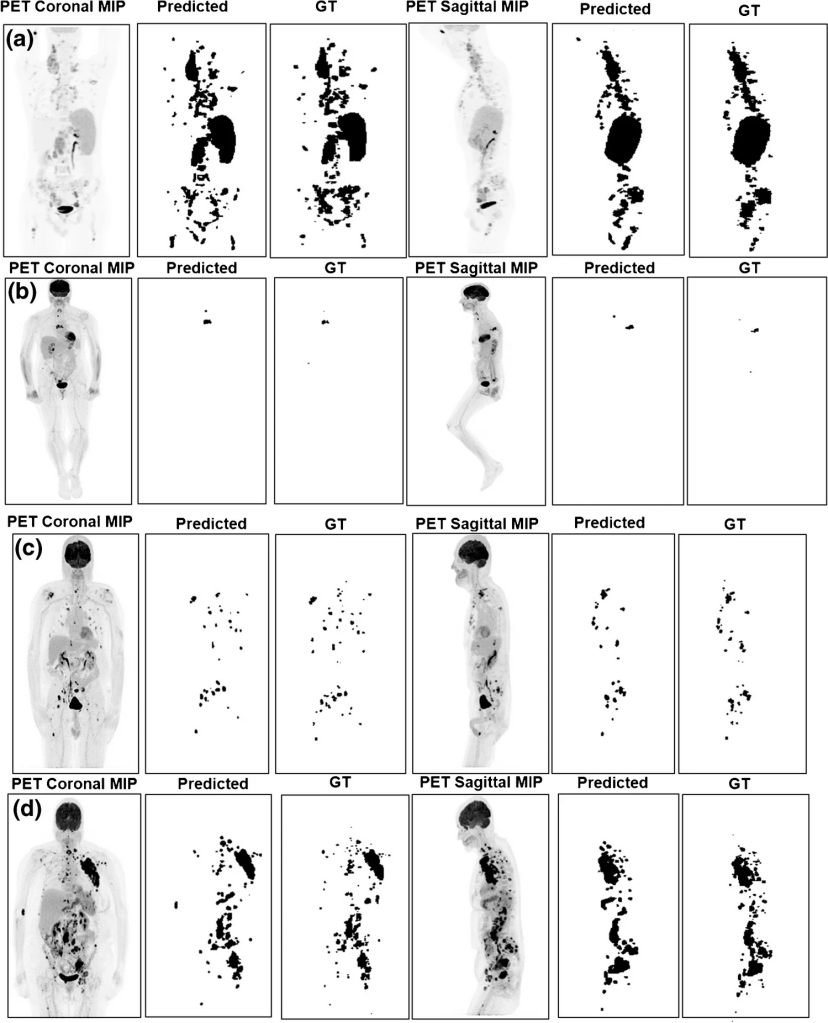
Example results of the segmented TMTV on data from UW center. a Hodgkin case, DSC = 0.83, TMTV relative error = 0.18. b DLBCL case, DSC = 0.66, TMTV relative error = 0.10. c Hodgkins case, DSC = 0.76. d DLBCL, DSC = 0.67
Claim: TMTV-Net represents a pivotal advancement in the field of tumor segmentation using 2-[18F] FDG PET/CT scans, offering a unique set of capabilities that contribute to superior generalizability when compared to existing approaches. Its integration of deep supervision within the framework of 3D U-Nets, coupled with the implementation of multi-resolution techniques, plays a pivotal role in mitigating the challenges posed by dataset shift. This strategic combination not only enhances the precision of tumor segmentation but also ensures the model reliability across diverse datasets. TMTV-Net demonstrates strong performance and generalizability in TMTV segmentation across multi-site external datasets, encompassing various lymphoma subtypes. A negligible reduction of 2% in overall performance during testing on external data highlights robust model generalizability across different centers and cancer types, likely attributable to its training with resampled inputs.
Remarks: TMTV-Net is publicly available, allowing easy multi-site evaluation and generalizability analysis on datasets from different institutions. A modified version of 3D U-Nets was used as segmentation model incorporating a deep supervision architecture [Ext. Ref]. Deeply supervised 3D U-Net uses additional supervision by making predictions at intermediate decoder layers as well. These intermediate predictions are then combined during training to compute the final loss, which aids in alleviating the vanishing gradient problem and facilitates faster convergence during training. A deep supervision module was used to apply deep supervision to intermediate layers and combine their outputs to compute the final loss. A semi-supervised loss function was used, composed of cross-entropy (CE) and Dice loss as supervised losses, and an unsupervised boundary-based loss term, namely, Mumford-Shah (MS) [Ext. Ref., Ext. Ref.]

wherein y is the output of the network, g is the ground truth, and θ is the network parameter [Ext. Ref.].
While a significant role was observed for the Mumford-Shah term in enhancing overall robustness particularly with limited datasets, in the current study, TMTV-net does not involve scenarios with scarce labeled data, and consequently, the weight assigned to this term is relatively low. The tuned parameters were:
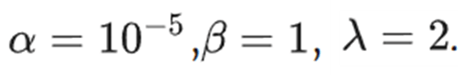
Test Time Augnmentation (TTA) involves generating multiple transformed copies of a test input and integrating the predictions obtained from these augmented images. By adopting TTA and considering the predictions from augmented images in addition to the “clean” images from the testing dataset, a more accurate final prediction can be achieved. This process usually involves averaging the predictions of each image, and it may also incorporate learnable weights to form a weighted average, ultimately contributing to superior segmentation outcomes and performance evaluation for various applications. The schematic of the TTA approach is shown below. TTA encompasses four steps: augmentation, prediction, inverse transform, and aggregation.
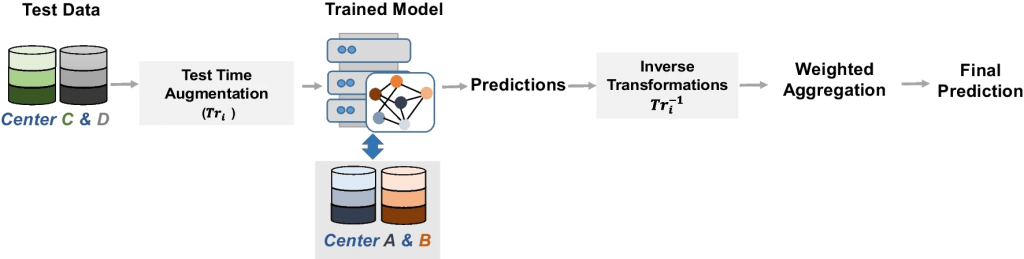
The adopted test time augmentation approach, involves the training of the model on data from center A and B. Test data includes data from centers C and D
To evaluate TMTV-Net on classical Hodgkin lymphoma cases (n = 30), in addition to DSC, qualitative analysis was conducted. For comparison to SOTA approaches, frameworks were considered based on nnU-Net as deployed by Blanc-Durand et al. [Ext. Ref.], deep evidential network by Huang et al. [Ext. Ref.], and Swin UNETR by Hatamizadeh et al. [Ext. Ref.] trained and tested on same data. An ablation analysis was performed on the cascaded segmentation approach to examine the key components of our proposed method. Specifically, the developers applied (i) a baseline approach using single-resolution 3D U-Net and (ii) only the first step (without cascaded refinement).
Future work could include the development of a user interface for active learning, allowing physicians to be more involved in segmentation. In addition, the addition of a convolutional layer is investigated to automatically learn the best possible combination of multiple-resolution models.
Reference Publication (to be cited if model is used for research purposes)
License: TMTV-Net is shared for research-use only. COMMERCIAL USE IS PROHIBITED for the time being. For further information please email: frizi@bccrc.ca