Task: Automatic organ and tumor segmentation for nanomedicine pharmacokinetic studies.
Description: NanoMASK model is a 3D U-Net adapted deep learning tool capable of rapid, automatic organ segmentation of multimodal imaging data that can output key clinical dosimetry metrics without manual intervention. NanoMASK is a dep learning application based on the nnUNet framework and adapted to a multifaceted database of longitudinal PET/CT whole-body scans of mice.
Contributions of NanoMask’s reference publication:
- demonstration of NanoMASK’s ability to generate highly accurate, automated, three-dimensional contours of multiple organ systems relative to the manually contoured ground truth.
- extraction of important pharmacokinetic measures for the functional imaging data that correlated highly with the values extracted from manual data processing.
- exploration of the dependencies of the NanoMASK model on various dimensions of this dataset, including modality, imaging timepoint, and tumor status, to highlight the importance of training on a nanomaterial-centric dataset with varied functional imaging contrast and its implications for auto-segmentation accuracy.
- validation of the model’s generalizability through application to external datasets with different nanoparticles, experimental timeframes, and imaging systems
Validation: The model was trained on 355 manually-contoured paired PET/CT data volumes of healthy or 4T1 orthotopic breast tumor-bearing mice injected with a variety of nanomaterials (a variety of different lipid-shelled microbubbles, agents which exhibit pharmacokinetic profiles similar to lipid nanoparticles) and imaged over 48 hours post injection.
Results: NanoMASK produced 3-dimensional contours of the heart, lungs, liver, spleen, kidneys, and tumor with high volumetric accuracy (pan-organ average %DSC of 92.5). NanoMASK’s auto-segmentation performed very well following 5-fold cross validation. Machine-generated contours were easily visualized alongside the base PET/CT data and appeared virtually indistinguishable when viewed next to the ground-truth contours as illustrated below. When quantitatively assessed, machine-generated contours displayed high spatial overlap with ground-truth contours for all organs tested.
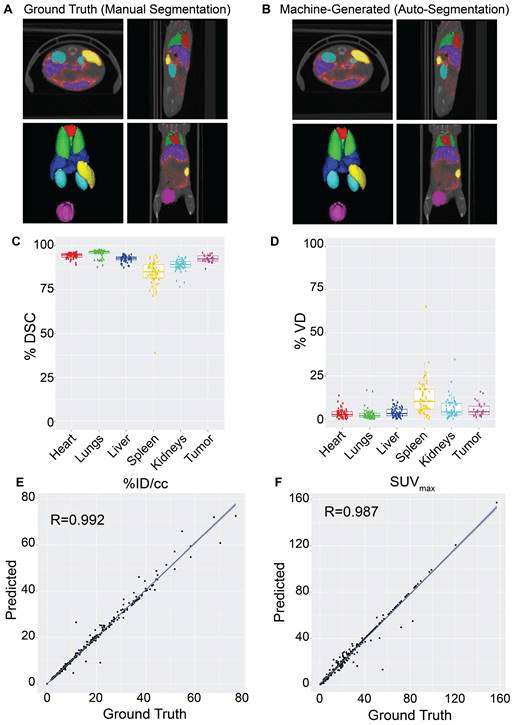
Pharmacokinetic metrics including %ID/cc, %ID, and SUVmax achieved correlation coefficients exceeding R = 0.987 and relative mean errors below 0.2%. NanoMASK was applied to novel datasets of lipid nanoparticles and antibody-drug conjugates with a minimal drop in accuracy, illustrating its generalizability to different classes of nanomedicines. Furthermore, 20 additional auto-segmentation models were developed using training data subsets based on image modality, experimental imaging timepoint, and tumor status. These were used to explore the fundamental biases and dependencies of auto-segmentation models built on a 3D U-Net architecture, revealing significant differential impacts on organ segmentation accuracy.
Claims: NanoMASK is an easy-to-use, adaptable deep learning network tool supporting automatic organ and tumor segmentation and designed for improving accuracy and throughput in imaging-based pharmacokinetic studies of nanomedicine. According to its developers, NanoMASK is the first auto-segmentation tool developed specifically for applications in nanomedicine. It combines both anatomical and functional imaging data to produce high quality contours of key organ systems related to agent pharmacokinetics and biodistribution. It was shown to be highly robust across different qualities of input data and generalizable to several nanomedicine classes. Importantly, it can generate pharmacokinetic outputs automatically with extremely high accuracy relative to manually calculated data. This poses to significantly reduce the time and expertise required to utilize nanomedicine preclinical imaging data to its fullest potential.
The model has been made publicly available to all readers for automatic segmentation and pharmacokinetic analysis across a diverse array of nanoparticles, expediting agent development. The open-access usage of the model or its principal architecture may facilitate its integration into the preclinical pipeline for nanomedicine platform optimization and expedite its more laborious aspects.
