Task: Automated Brain Tumor Detection and Segmentation for Treatment Response Assessment Using Amino Acid PET
Description: JuST_BrainPET: Juelich Segmentation Tool for Brain Tumor PET is a model trained for metabolic tumor volume (MTV) segmentation in amino acid PET scans of brain tumor patients using the tracer O-(2-[18F]fluoroethyl)-L-tyrosine (18F-FET).
The JuST_BrainPET model is based on the self-configuring nnU-Net v1 framework as a pretrained model for automated 3-dimensional MTV segmentation of brain tumors in amino acid 18F-FET exams. It has been developed by the Institute of Neuroscience and Medicine, Forschungszentrum Juelich GmbH in Germany and was first introduced in a reference study where its performance was evaluated in terms of response assessment in patients with gliomas.
In the JuST_BrainPET model’s reference study, the nnU-Net v1 was used, which is a fully automated self-configuring artificial neural network for semantic segmentation. Starting from a dataset and the underlying annotations, nnU-Net performs an automated analysis of the training cases and subsequently configures the entire segmentation pipeline. The concept of nnU-Net v1 involves fixed parameters, heuristic rules dependent on the dataset, and empirically determined parameters. The nnU-Net v1 model is publicly available at GitHub. A full description and further details about nnU-Net can be found in that model’s reference publication.
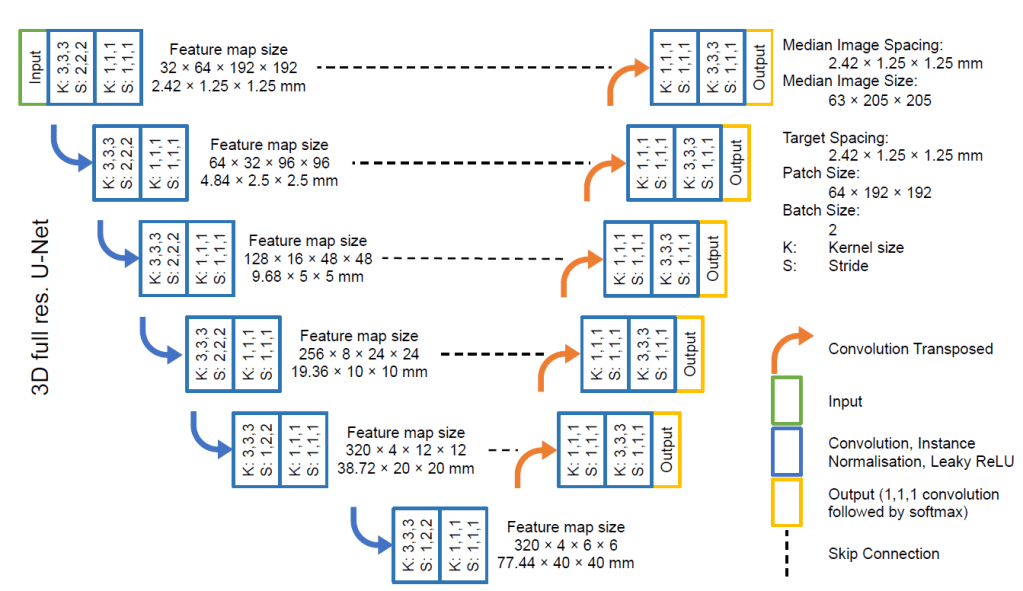
In JuST_BrainPET model’s reference study, the ‘3d_fullres’ configuration of the nnU-Net v1 was employed. The patch size was set to 64x192x192 mm3, and all images were resampled to a voxel size of 2.42×1.25×1.25 mm3. Image normalization was performed with z-score normalization. The batch size during training was chosen to be two. A detailed illustration of the resulting network architecture is shown below.
Evaluation: In total, 699 amino acid 18F-FET PET scans from 555 brain tumor patients at initial diagnosis or during follow-up were retrospectively evaluated (mainly glioma patients, 76%). 18F-FET PET MTVs were segmented semiautomatically by experienced readers. An artificial neural network (nnU-Net v1) was configured on 476 scans from 399 patients, and the network performance was evaluated on a test dataset including 223 scans from 156 patients. Surface and volumetric Dice similarity coefficients (DSCs) were used to evaluate segmentation quality. Finally, the network was applied to a recently published 18F-FET PET study on response assessment in glioblastoma patients treated with adjuvant temozolomide chemotherapy for a fully automated response assessment in comparison to an experienced physician.
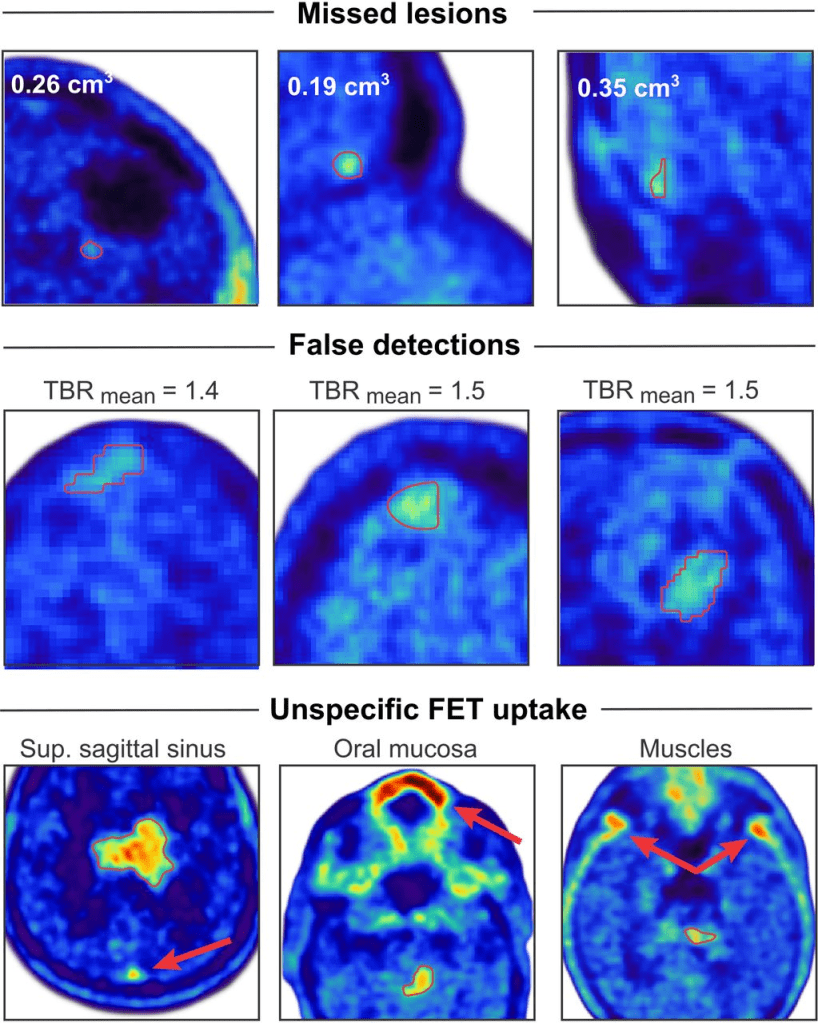
Results: In the test dataset, 92% of lesions with increased uptake (n = 189) and 85% of lesions with iso- or hypometabolic uptake (n = 33) were correctly identified. Importantly, none of the anatomic regions that showed a physiologically increased uptake, such as in the superior sagittal sinus, were considered to be tumors by the network. This resulted in a mean F1 score of 92%, a sensitivity of 93%, and a positive predictive value of 95% for lesion detection. Patient examples showing lesions missed by the network, false detections, and examples of regions showing physiologically increased uptake are provided in the figure below:
Single lesions with a contiguous uptake had the highest DSC, followed by lesions with heterogeneous, noncontiguous uptake and multifocal lesions (surface DSC: 0.96, 0.93, and 0.81 respectively; volume DSC: 0.83, 0.77, and 0.67, respectively). Change in MTV, as detected by the automated segmentation, was a significant determinant of disease-free and overall survival, in agreement with the physician’s assessment.
Claim: The deep learning–based automated MTV segmentation of brain tumors in 18F-FET PET images as implemented with the Just_BrainPET model allows reliable, robust, and fully automated evaluation of MTV in brain tumor patients and demonstrates clinical value for automated response assessment.
Data Sharing Statement: The dataset, including 18F-FET PET image data and segmentations, is available on request. In addition, data analysis scripts in Python are available on request. The trained network model (JuST_BrainPET) is available on GitHub.
