Task: Semi-automated tissue-fraction estimation-based lesion segmentation from human PET images for quantitative assessments of oncologic diseases.
Description: The proposed model implements a novel Bayesian approach to tissue-fraction estimation (TFE) for oncological PET segmentation. Specifically, the segmentation problem is posed as a task of estimating the fractional volume that the tumor occupies within each voxel of an image. The proposed Bayesian approach estimates the posterior mean of the fractional tumor volume at each voxel. Conventional segmentation methods are typically classification-based, i.e. classifying each voxel in the image as belonging to a certain tissue class. Thus, these methods are inherently limited in modeling TFEs. While probabilistic techniques can provide estimates of probabilities that each image voxel belongs to a tissue class, these probabilistic estimates are unrelated to TFEs. The developers of this model address this inherent limitation by framing the segmentation task as an estimation problem, where the fractional volume that the tumor occupies in each voxel is estimated. Through this strategy, explicit modeling of the TFEs became possible while performing segmentation.
The objective of this optimization problem is to design a model that can estimate the TFEs from a reconstructed image at each voxel. For that purpose, a cost function must first be defined that penalizes deviation of TFE estimates from the actual true fractional volume that the tumor occupies within each voxel. A common cost function is the ensemble mean squared error (EMSE), which is the mean squared error averaged over noise realizations and the true fraction values. However, the fractional volume estimates, can be constrained to lie within the [0,1] range, and the EMSE loss does not directly incorporate this constraint. In contrast, using the binary cross-entropy (BCE) loss as the penalizer allows the incorporation of this constraint on the TFE estimates directly, as previously suggested in Creswell et al. 2017.
Implementation: The method was implemented and evaluated on a simplified per-slice basis. Thus, for each pixel in the 2D reconstructed image, the optimizer was designed to yield the posterior mean estimate of the true tumor-fraction area (TFA), The procedure to implement this optimizer is then described.
Estimating the posterior mean estimate requires sampling from its respective posterior probability distribution. Sampling from this distribution is challenging as this distribution is high-dimensional and does not have a known analytical form. To address this issue, the proposed method was implemented using a supervised learning-based approach. Specifically, an encoder–decoder network was constructed, as shown in the figure below.
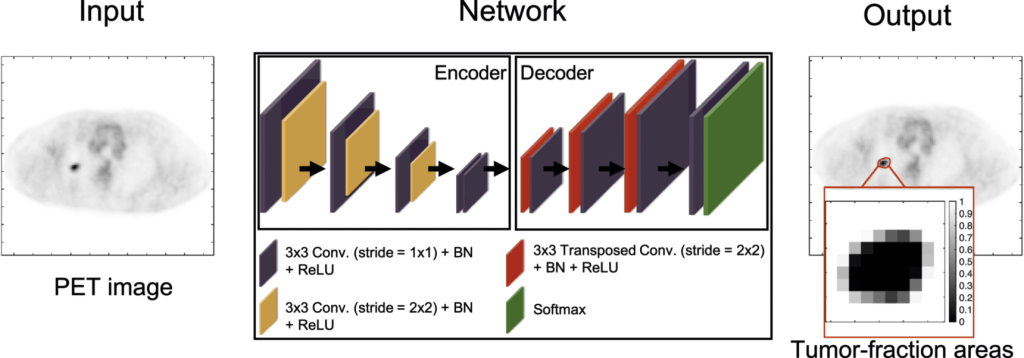
During the training phase, this network is provided with a population of PET images, and the corresponding ground-truth TFA map, i.e. a vector for each image. The network can then be trained to yield the posterior-mean estimate given the input PET image. This learning process is attained by minimizing the BCE cost function over this population of images.
The network architecture is similar to those for estimation tasks, such as image denoising (Creswell et al 2017) and image reconstruction (Nath et al 2020). The network is partitioned into a contracting and an expansive path. The contracting path learns the spatial information from the input PET images and the expansive path maps the learned information to the estimated TFA map for each input image. Skip connections with element-wise addition were applied to feed the features extracted in the contracting path into the expansive path to stabilize the training and improve the learning performance (Mao et al 2016). In the final layer, the network yields the estimate of the TFAs. A detailed description of the network architecture is provided in the Table below:
| Layer type | Filter size | # of filters | Stride | Input size | Output size | |
|---|---|---|---|---|---|---|
| Layer 1 | Conv. | 3 × 3 | 32 | 1 × 1 | 168 × 168 × 1 | 168 × 168 × 32 |
| Layer 2 | Conv. | 3 × 3 | 32 | 2 × 2 | 168 × 168 × 32 | 84 × 84 × 32 |
| Layer 3 | Conv. | 3 × 3 | 64 | 1 × 1 | 84 × 84 × 32 | 84 × 84 × 64 |
| Layer 4 | Conv. | 3 × 3 | 64 | 2 × 2 | 84 × 84 × 64 | 42 × 42 × 64 |
| Layer 5 | Conv. | 3 × 3 | 128 | 1 × 1 | 42 × 42 × 64 | 42 × 42 × 128 |
| Layer 6 | Conv. | 3 × 3 | 128 | 2 × 2 | 42 × 42 × 128 | 21 × 21 × 128 |
| Layer 7 | Conv. | 3 × 3 | 256 | 1 × 1 | 21 × 21 × 128 | 21 × 21 × 256 |
| Layer 8 | Conv. | 3 × 3 | 256 | 1 × 1 | 21 × 21 × 256 | 21 × 21 × 256 |
| Layer 9 | Transposed Conv. | 3 × 3 | 128 | 2 × 2 | 21 × 21 × 256 | 42 × 42 × 128 |
| Layer 9 | Skip connection (add layer 5) | — | — | — | 42 × 42 × 128 | 42 × 42 × 128 |
| Layer 10 | Conv. | 3 × 3 | 128 | 1 × 1 | 42 × 42 × 128 | 42 × 42 × 128 |
| Layer 11 | Transposed Conv. | 3 × 3 | 64 | 2 × 2 | 42 × 42 × 128 | 84 × 84 × 64 |
| Layer 11 | Skip connection (add layer 3) | — | — | — | 84 × 84 × 64 | 84 × 84 × 64 |
| Layer 12 | Conv. | 3 × 3 | 64 | 1 × 1 | 84 × 84 × 64 | 84 × 84 × 64 |
| Layer 13 | Transposed Conv. | 3 × 3 | 32 | 2 × 2 | 84 × 84 × 64 | 168 × 168 × 32 |
| Layer 13 | Skip connection (add layer 1) | — | — | — | 168 × 168 × 32 | 168 × 168 × 32 |
| Layer 14 | Conv. | 3 × 3 | 32 | 1 × 1 | 168 × 168 × 32 | 168 × 168 × 32 |
| Layer 15 | Conv. | 3 × 3 | 2 | 1 × 1 | 168 × 168 × 32 | 168 × 168 × 2 |
| Output | Softmax | — | — | — | 168 × 168 × 2 | 168 × 168 × 2 |
The goal of the model’s implementation is to explicitly model the TFEs while performing tumor segmentation. Thus, the training strategy and network architecture were specifically designed for this goal by defining the ground truth as the TFAs for each image. This is in contrast to the conventional DL-based segmentation methods, where, in the ground truth, each pixel is exclusively assigned to the tumor or the normal tissue class and the network is trained to classify each pixel as either tumor or background. Further, while the conventional DL-based methods can output a probabilistic estimate for each image pixel, this estimate is only a measure of classification uncertainty, and thus has no relation to TFEs, unlike the proposed method.
The network was trained via the Adam optimization algorithm (Kingma and Ba 2014). In the various experiments mentioned later, the network hyperparameters were optimized on a training set via five-fold cross validation. The network training was implemented in Python 3.6.9, Tensorflow 1.14.0, and Keras 2.2.4. Experiments were performed on a Linux operating system with two NVIDIA Titan RTX graphics processing unit cards.
Evaluation: The model was first evaluated using clinically realistic 2D simulation studies with known ground truth, in the context of segmenting the primary tumor in PET images of patients with lung cancer. Evaluation was then performed on clinical images of patients with stage IIB/III non-small cell lung cancer from the ACRIN 6668/RTOG 0235 multi-center clinical trial.
Since the proposed method is an estimation-based segmentation approach, the evaluation study employed performance metrics for both the task of estimating the true TFA map and of segmenting the tumor.
Performance on the estimation task was evaluated using the EMSE between the true and estimated TFA maps. EMSE provides a combined measure of bias and variance over the distribution of true values and noise realizations, and is thus considered as a comprehensive figure of merit for estimation tasks (Barrett and Myers 2013). The error in the estimate of the TFA maps and the tumor area was quantified using the pixel-wise EMSE and normalized area EMSE, respectively. The proposed evaluation method estimates the TFA within each pixel, which is a continuous-valued output.
For the evaluation of segmentation methods that yield such non-binary output, as in Taha and Hanbury 2015, the spatial-overlap-based metrics can be derived based on the four cardinalities of confusion matrix, namely the true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN). Furthermore, the spatial-overlap metric of Dice similarity coefficient (DSC) and Jaccard similarity coefficient (JSC) were used to measure the agreement between the true and estimated segmentation.
The performance of the proposed method was also qualitatively evaluated on the task of estimating the TFA map. For this purpose, ground-truth and estimated tumor topographic maps were first constructed from the true and estimated TFA maps using the contour function in MATLAB (MathWorks, Natick, Mass).
Results: The simulated evaluation studies demonstrated that the method accurately estimated the tumor-fraction areas and significantly outperformed widely used conventional PET segmentation methods, including a U-net-based method, on the task of segmenting the tumor. In addition, the proposed method was relatively insensitive to PVEs and yielded reliable tumor segmentation for different clinical-scanner configurations.
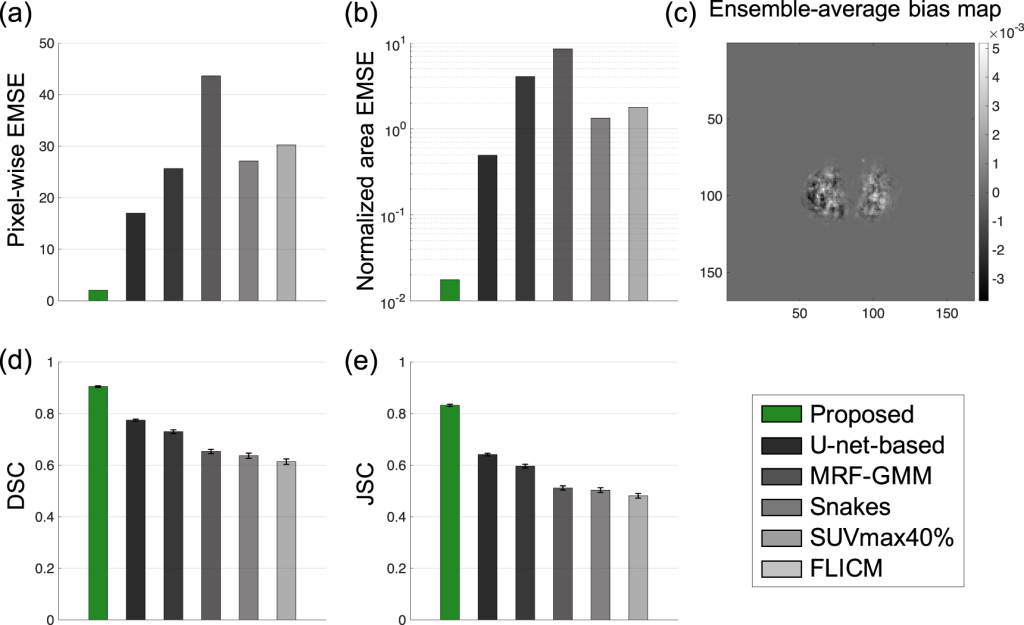
The experimental evaluation results showed that the proposed method significantly outperformed all other considered methods and yielded accurate tumor segmentation on patient images with Dice similarity coefficient (DSC) of 0.82 (95% CI: 0.78, 0.86). In particular, the method accurately segmented relatively small tumors, yielding a high DSC of 0.77 for the smallest segmented cross-section of 1.30 cm2.
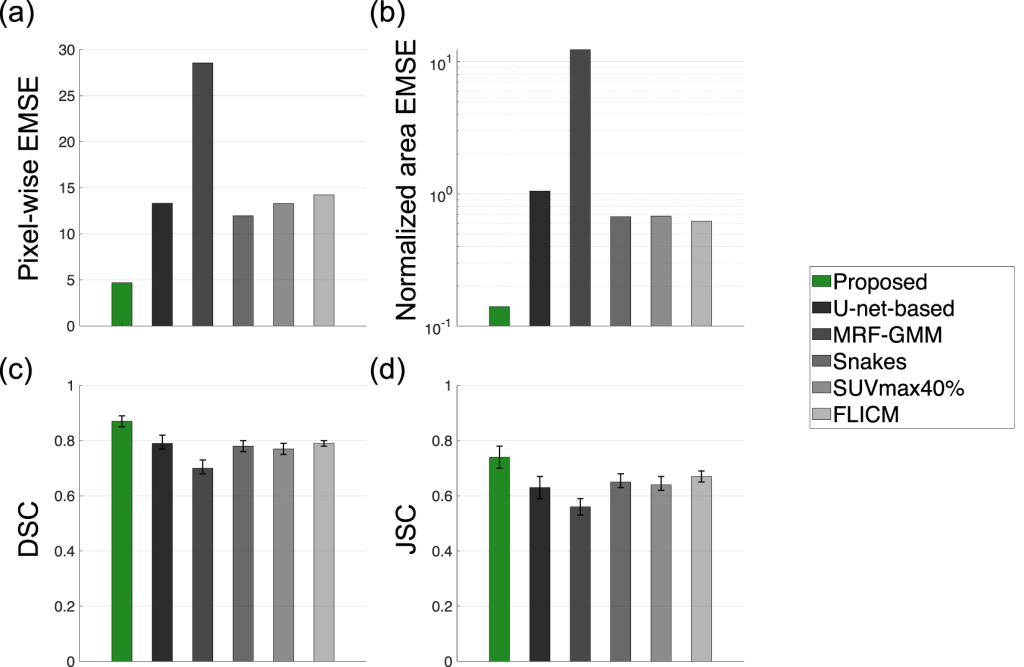
Limitations: The model’s reference study has acknowledged certain limitations associated with the proposed model. First, while the theory of the proposed method was developed in the context of 3D imaging, the evaluation studies were conducted on a per-slice basis to increase the size of training data and reduce the computational cost (Leung et al. 2020). However, implementing the method to 3D segmentation is relatively straightforward and would require only slight modifications to the network architecture, such as the ability to be input 3D images and output 3D tumor-fraction volume maps. Thus, the 2D convolutional layers in the network would be replaced by 3D convolutional layers. The overall network design would remain similar. The results shown in this study and a reference 3D SPECT segmentation study suggest that the proposed method will yield reliable performance for 3D tumor segmentation in PET, and this is an area of further research.
Additionally, in this study, the proposed method was used to segment the image into only two regions. However, the method is general, and in an ongoing study of 3D SPECT segmentation, this model was suucessfully applied to segment the images into seven different regions.
Another limitation is that the evaluation studies currently consider cases where only the primary tumor is present in an image. However, again, the method could be generalized to potentially segment multiple tumors present in the same image slice. Confirming this though would require additional evaluation studies.
Further, respiratory motion of the lung, which may also cause blurring of the tumor mask, was not considered in the proposed method. Extending the method to account for lung motion is also an important research area.
Finally, the method does not incorporate tumor information from CT images while segmenting PET images. Incorporating information from CT images can provide a prior distribution of the TFAs for the estimation task. Thus, investigating the incorporation of CT images into the proposed method is another important research direction.
Claim: The evaluation study published by the model developers demonstrated the efficacy of the proposed model to accurately segment tumors in PET images.
GitHub Pages: Open-source code for the proposed tissue-fraction estimation model for oncological PET segmentation
