Task: Automatic Impression Generation for PET Reports Using Large Language Models (LLMs)
Description: A collection of twelve open-source language models fine-tuned on a corpus of 37,370 retrospective PET reports were collected from University of Wisconsin, Madison, WI, USA between 2010 and 2022. All models were trained using the teacher-forcing algorithm, with the report findings and patient information as input and the original clinical impressions as reference. An extra input token encoded the reading physician’s identity, allowing models to learn physician-specific reporting styles.
Models Comparative Evaluation: To compare the performances of different LLMs, various automatic 30 evaluation metrics were computed and benchmarked against physician preferences. The winner LLM’s impressions were then evaluated for their clinical utility, against original clinical impressions, by three nuclear medicine physicians across 6 quality dimensions (3-point scales) and an overall utility score (5-point scale). Each physician reviewed 12 of their own reports and 12 reports from other physicians.
The 12 evaluated fine-tuned LLMs can be found below:
- BERT2BERT-PET
- BART-PET
- BioBART-PET
- PEGASUS-PET
- T5v1.1-PET
- Clinical-T5-PET
- Flan-T5-PET
- GPT2-XL-PET
- OPT-1.3B-PET
- LLaMA-LoRA-PET
- Alpaca-LoRA-PET
The categories of all 30 evaluation metrics employed for the LLM performance comparison are summarized below:
| Category | Definition | Corresponding Evaluation Metrics |
|---|---|---|
| Lexical overlap-based metrics | These metrics measure the overlap between the generated text and the reference in terms of textual units, such as n-grams or word sequences | ROUGE-1, ROUGE-2, ROUGE-3, ROUGE-L, ROUGE-LSUM, BLEU, CHRF, METEOR, CIDEr |
| Embedding-based metrics | These metrics measure the semantic similarity between the generated and reference texts using pretrained embeddings | ROUGE-WE-1, ROUGE-WE-2, ROUGE-WE-3, BERTScore, MoverScore |
| Graph-based metrics | These metrics construct graphs using entities and their relations extracted from the sentences, and evaluate the summary based on these graphs | RadGraph |
| Text generation-based metrics | These metrics assess the quality of generated text by framing it as a text generation task using sequence-to-sequence language models | BARTScore, BARTScore + PET PEGASUSScore + PET, T5Score + PET, PRISM |
| Supervised regression-based metrics | These metrics require human annotations to train a parametrized regression model to predict human judgments for the given text | S3-pyr, S3-resp |
| Question answering-based metrics | These metrics formulate the evaluation process as a question-answering task by guiding the model with various questions | UniEval |
| Reference-free metrics | These metrics do not require the reference text to assess the quality of the generated text. Instead, they compare the generated text against the source document | SummaQA, BLANC, SUPERT, Stats-compression, Stats-coverage, Stats-density, Stats-novel trigram |
The implementation methods employed by the comparative evaluation study are shared on GitHub:
- fastAI Implementation: simple and easy to use
- Non-trainer Implementation: more flexible
- Trainer (with deepspeed) Implementation: reduce memory usage and accelerate training
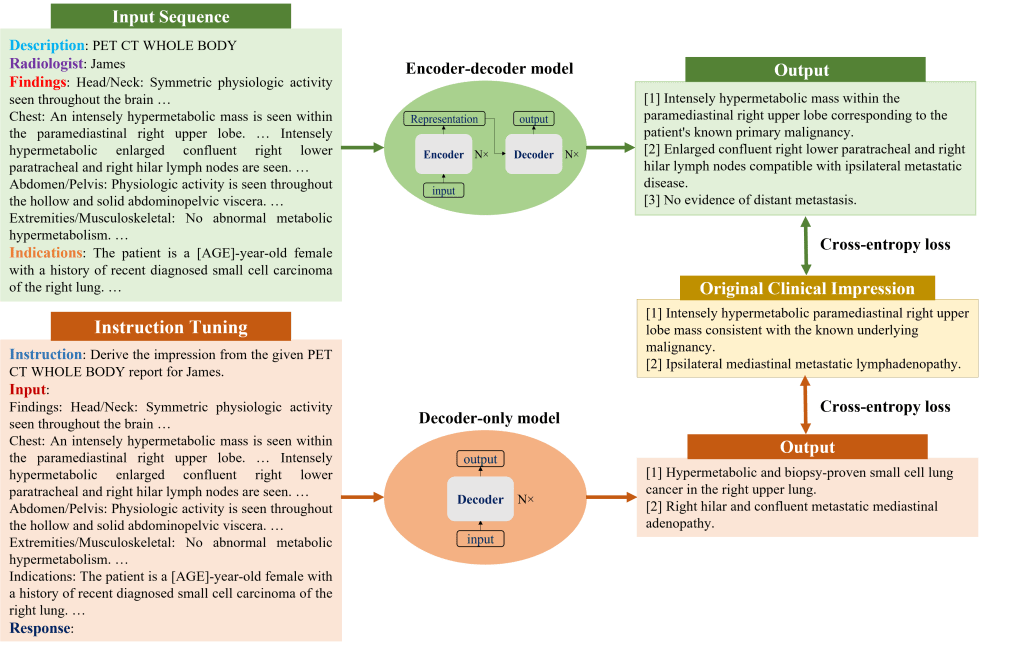
Both encoder-decoder and decoder-only language models were evaluated. Considering their different architectures, input templates were customized as illustrated in below figure. For encoder-decoder models, the first lines describe the categories of PET scans, while the second lines encode each reading physician’s identity using an identifier token.
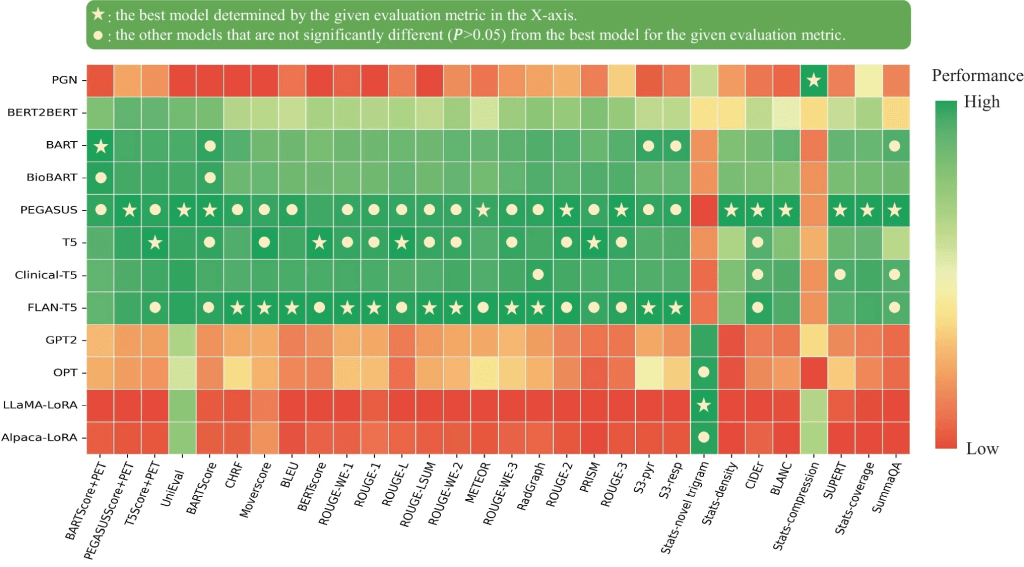
Results: On average, the PEGASUS LLM quantitative evaluation scores outperformed all other LLMs. When physicians assessed PEGASUS LLM impressions generated in their own style, 89% were considered clinically acceptable, with a mean utility score of 4.08/5. On average, physicians rated these personalized impressions as comparable in overall utility to the impressions dictated by other physicians (4.03, P = 0.41). In summary, the comparative evaluation study demonstrated that personalized impressions generated by PEGASUS LLM were clinically useful in most cases, highlighting its potential to expedite PET reporting by automatically drafting impressions.
Claim: The fine-tuned large language model provides clinically useful, personalized impressions based on PET findings. To the best of the authors’ knowledge, this is the first attempt to automate impression generation for whole-body PET reports.
Key Points (from GitHub documentation comparing the 12 fine-tuned LLMs):
- 📈 Among 30 evaluation metrics, domain-adapted BARTScore and PEGASUSScore exhibited the highest correlations (Spearman’s ρ correlation=0.568 and 0.563) with physician preferences, yet they did not reach the level of inter-reader correlation (ρ=0.654).
- 🏆 Of all fine-tuned large language models, encoder-decoder models outperformed decoder-only models, with PEGASUS emerging as the top-performing model.
- 🏅 In the reader study, three nuclear medicine physicians considered the overall utility of personalized PEGASUS-generated impressions to be comparable to clinical impressions dictated by other physicians.
PEGASUS-PET was fine-tuned based on Google’s PEGASUS LLM implementation.
Data Availability Statement: The radiology reports used in this study are not publicly available due to privacy concerns related to HIPAA. However, upon reasonable request and approval of a data use agreement, they can be made available for research purposes. COG AHOD1331 clinical trial data is archived in NCTN Data Archive.
Documentation: Usage, PET Human Experts Report Evaluation
