Task: Fully automated tissue-fraction estimation-based segmentation of the caudate, putamen, and Globus Pallidus (GP) from human brain DaT-SPECT images for quantitative assessments of Parkinson’s Disease.
Description: The DaT-SPECT-Segmentation model estimates the posterior mean of the fractional volumes occupied by the caudate, putamen, and GP within each voxel of a three-dimensional DaT-SPECT image. The estimate is obtained by minimizing a cost function based on the binary cross-entropy loss between the true and estimated fractional volumes over a population of SPECT images, where the distribution of true fractional volumes is obtained from existing populations of clinical magnetic resonance images. The method is implemented using a supervised deep-learning-based approach.
As elaborated in the model’s reference publication, the first major contribution of the model is to advance the idea of estimation-based segmentation, initially proposed by Liu et al. as a semi-automated segmentation method from 2D oncological PET images, to develop a Bayesian tissue-fraction estimation (TFE) method for fully automated and simultaneous segmentation of the caudate, putamen, and GP from 3D DaT-SPECT images. The proposed model provides a mechanism to obtain the true fractional volumes of the considered regions from other imaging modalities, thus not requiring manual segmentation of the SPECT images for obtaining the ground truth.
The second major contribution of the model is the proposal of a novel strategy to objectively evaluate segmentation methods on the task of regional uptake quantification. In DaT SPECT, segmentation is performed for the clinical task of quantifying the mean uptake in the various regions of the brain. Thus, segmentation methods should be evaluated based on this quantification task. Towards this goal, the model developers proposed a projection-domain-quantification-based strategy to perform such objective task-based evaluation in an optimal manner.
In the model’s reference study, the the MR-defined fractional volumes is considered as the surrogate for the ground truth assuming perfect 3D co-registration between brain MR and DaT-SPECT images. The objective of the model is to estimate the true fractional volumes from the input SPECT image.
Moreover, the binary cross-entropy (BCE) loss was selected as the basis of the cost function to be minimized since this loss term automatically incorporates the constraint that the tissue fraction volume estimates must always lie between 0 and 1 by definition.
The proposed method was implemented using a supervised DL-based approach to overcome the challenges of sampling from the posterior distribution of the tissue fraction estimated volumes due to the high dimensionality and unknown analytical form of this distribution. Specifically, a standard encoder–decoder-based neural network was constructed. During training, the network was input a population of 3D SPECT images and the corresponding true fractional volumes occupied by the caudate, putamen, GP, and background. The network was then trained to minimize the BCE cost function using the Adam algorithm to yield, given an input DaT-SPECT image, the posterior-mean estimate of those true fractional volumes within each voxel of that SPECT image.
The architecture of the network is similar to that of networks designed for estimation tasks, such as image denoising and reconstruction. More specifically, the network comprises an encoder and a decoder. The encoder extracts local spatial features from the input SPECT image and the decoder maps the extracted features to the fractional volumes. In the final layer, the network outputs the fractional volumes occupied by the left and right caudate, putamen, and GP, and the background, within each voxel of the input SPECT image. To stabilize the network training, skip connections with element-wise addition were applied between the output of layers in the encoder and decoder. Additionally, dropout was applied to prevent overfitting. Further, instead of performing slice-by-slice or patch-based training, the network was designed to be input the whole 3D image to learn the maximal amount of global contextual information. Implementation of the proposed method is illustrated in the figure below,
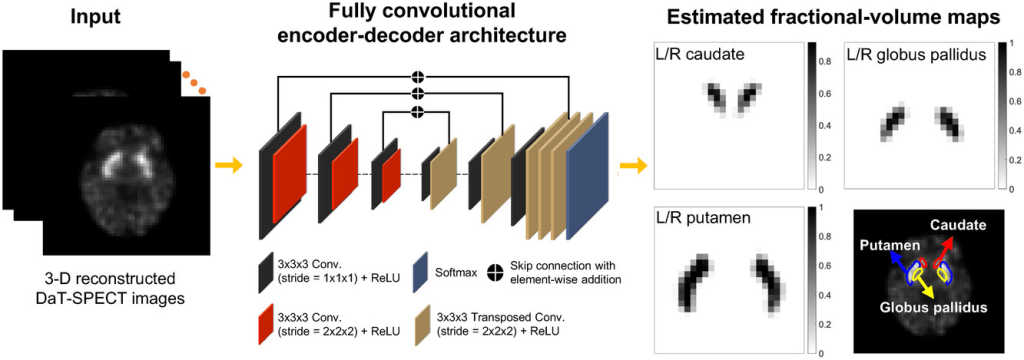
A more detailed description of the network architecture is provided in the table below.
| Layer | Layer type | Number of filters | Filter size | Stride size | Input size | Output size |
|---|---|---|---|---|---|---|
| 1 | Conv. | 32 | 3× 3× 3 | 1× 1× 1 | 128× 128× 128× 1 | 128× 128× 128× 32 |
| 2 | Conv. | 32 | 3× 3× 3 | 2× 2× 2 | 128× 128× 128× 32 | 64× 64× 64× 32 |
| 3 | Conv. | 64 | 3× 3× 3 | 1× 1× 1 | 64× 64× 64× 32 | 64× 64× 64× 64 |
| 4 | Conv. | 64 | 3× 3× 3 | 2× 2× 2 | 64× 64× 64× 64 | 32× 32× 32× 64 |
| 5 | Conv. | 128 | 3× 3× 3 | 1× 1× 1 | 32× 32× 32× 64 | 32× 32× 32× 128 |
| 6 | Conv. | 128 | 3× 3× 3 | 2× 2× 2 | 32× 32× 32× 128 | 16× 16× 16× 128 |
| 7 | Conv. | 256 | 3× 3× 3 | 1× 1× 1 | 16× 16× 16× 128 | 16× 16× 16× 256 |
| 8 | Transposed Conv. | 128 | 3× 3× 3 | 2× 2× 2 | 16× 16× 16× 256 | 32× 32× 32× 128 |
| 9 | Add Layer 5 | – | – | – | 32× 32× 32× 128 | 32× 32× 32× 128 |
| 10 | Conv. | 128 | 3× 3× 3 | 1× 1× 1 | 32× 32× 32× 128 | 32× 32× 32× 128 |
| 11 | Transposed Conv. | 64 | 3× 3× 3 | 2× 2× 2 | 32× 32× 32× 128 | 64× 64× 64× 64 |
| 12 | Add Layer 3 | – | – | – | 64× 64× 64× 64 | 64× 64× 64× 64 |
| 13 | Conv. | 64 | 3× 3× 3 | 1× 1× 1 | 64× 64× 64× 64 | 64× 64× 64× 64 |
| 14 | Transposed Conv. | 32 | 3× 3× 3 | 2× 2× 2 | 64× 64× 64× 64 | 128× 128× 128× 32 |
| 15 | Add Layer 1 | – | – | – | 128× 128× 128× 32 | 128× 128× 128× 32 |
| 16 | Conv. | 32 | 3× 3× 3 | 1× 1× 1 | 128× 128× 128× 32 | 128× 128× 128× 32 |
| 17 | Conv. | 7 | 3× 3× 3 | 1× 1× 1 | 128× 128× 128× 32 | 128× 128× 128× 7 |
| Output | Softmax | – | – | – | 128× 128× 128× 7 | 128× 128× 128× 7 |
It should be noted that while training the proposed method, the ground truth is defined as the fractional volumes occupied by each region within each voxel of the input SPECT image. Thus, the method is specifically designed and trained to model the TFEs while performing segmentation. In contrast, in conventional DL-based segmentation methods (Leung et al., Lin et al.), the ground truth is defined such that each voxel is assigned as belonging to a specific region. Thus, these methods are inherently limited in modeling the TFEs. Further, while these methods can output a probabilistic estimate of each voxel belonging to a region, this probability is unrelated to the TFEs.
Evaluation: The proposed method was evaluated based on two criteria.
- assessment of the spatial overlap and shape similarity between the true and estimated segmentations of the caudate, putamen, and GP, and
- evaluation of the proposed method for the task of quantifying the DaT uptake within those segmented regions.
The description of this evaluation study is grouped into five subparts, namely:
- data collection,
- network training,
- testing procedure,
- process to extract task-specific information, and
- figures of merit.
This evaluation procedure is in accordance to a recently proposed framework for objective evaluation of AI-based methods for medical imaging:
- Reference Publication #1: Nuclear Medicine and Artificial Intelligence: Best Practices for Evaluation (the RELAINCE Guidelines)
- Reference Publication #2: Nuclear Medicine and Artificial Intelligence: Best Practices for Algorithm Development
- Reference Publication #3: Objective Task-Based Evaluation of Artificial Intelligence-Based Medical Imaging Methods: Framework, Strategies, and Role of the Physician
Data Collection:
the proposed method was evaluated using clinically guided highly realistic simulation studies.
A total of 580 T1-weighted MR images of cognitively healthy individuals were obtained from the OASIS-3 database (accessible online here). From these MR images, the caudate, putamen, and GP in both left and right hemispheres of the brain were segmented using the Freesurfer software. Segmentations were performed in the Montreal Neurological Institute (MNI) space and then transformed back into the native space of each patient, yielding 580 anatomical templates. The dimension of each template was 256×256×256, with a voxel size of 1mm×1mm×1mm. From each template, the supports of the regions of caudate, putamen, and GP were obtained. Clinically realistic DaT-tracer distributions within these regions were next simulated. For each template, the DaT-specific binding ratio (SBR) within each region was independently sampled from a Gaussian distribution with mean and standard deviation values obtained from clinical data. This sampling process helped to obtain a clinically realistic variability of regional DaT uptake in the simulated patient population. By the end of this process, a digital phantom population of 580 patients was obtained with anatomical templates derived directly from clinical MR images and DaT-tracer distributions guided by clinical data. The simulation process is illustrated in the figure below:
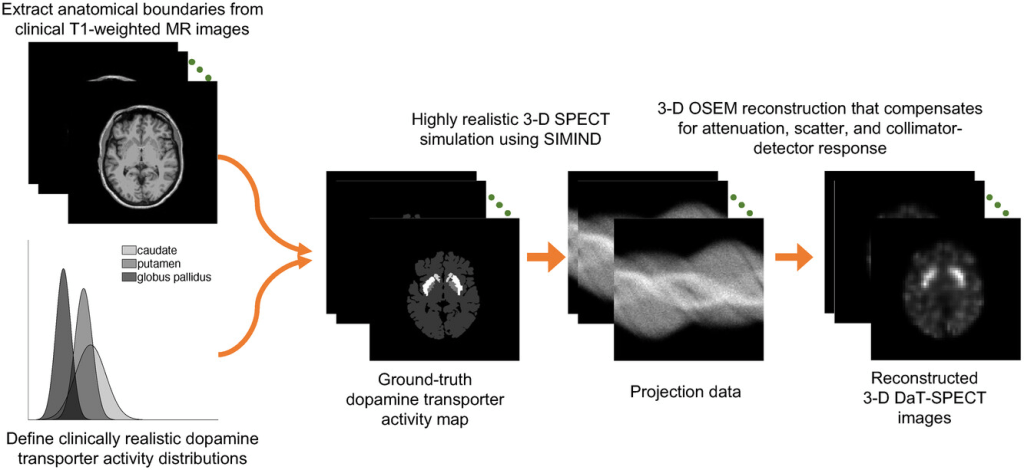
Network Training: Following the simulation procedure above, a total of 580 reconstructed 3D DaT-SPECT images were generated. Of these images, 480 were used for network training. The hyperparameters of the network were optimized via fivefold cross-validation. The remaining 100 images were reserved for evaluating the performance of the trained network.
The training strategy for the proposed model was designed to model the TFEs while performing segmentation. Thus, the ground truth was defined as the fractional volumes occupied by the caudate, putamen, GP, and background within each voxel of a SPECT image. For each patient, the Freesurfer-defined segmentations from the MR images provided the high-resolution support of each region. From this support, the true fractional volumes were obtained. The network was then trained by minimizing the BCE cost function, as referenced above.
Testing Procedure 1: In this study, the proposed method was evaluated based on spatial overlap and shape similarity between the true and estimated segmentations. Additionally, the proposed method was compared to several commonly used segmentation methods. Common methods for segmenting SPECT and PET images can be categorized into those based on thresholding, boundary detection, stochastic modeling, and learning. In this study, the following segmentation methods were comparatively evaluated:
- 40% SUV-max thresholding,
- Snakes,
- Markov random fields-Gaussian mixture model (MRF-GMM),
- U-net-based method
Testing Procedure 2: The goal of segmentation in DaT SPECT is to quantify the uptake within the segmented regions. Thus, an objective evaluation of a segmentation method should assess the efficacy of the method in performing this task (task-based evaluation). For this purpose, the model developers introduced a framework to objectively evaluate the proposed method for this particular quantification task. An important component of any task-based evaluation is the process to extract the task-specific information. The related details can be found in the model’s reference publication.
Figures of Merit: To quantify the spatial overlap between the true and estimated segmentations, it is recognized that for each voxel of a SPECT image, the proposed method yields continuous-valued estimates of fractional volumes. The Dice similarity coefficient (DSC) as proposed in Taha and Hanbury provides a figure of merit (FoM) to evaluate spatial overlap when segmentation methods yield such continuous-valued outputs. This FoM was thus used in this study. A higher value of DSC implies a more accurate segmentation performance.
In addition, the shape similarity between the true and estimated segmentations was quantified using the Hausdorff distance (HD). To compute the HD requires obtaining the isosurfaces from segmentations. For this purpose, the strategy described in Liu et al. was adopted.
The performance on the task of quantifying regional uptake was evaluated based on the accuracy and overall reliability of the estimated regional uptake over the 100 test patients, using the FoMs of ensemble normalized bias (NB) and ensemble normalized root mean square error (NRMSE), respectively.
Results: According to the model’s reference study, evaluations using clinically guided highly realistic simulation studies show that the DaT-SPECT-Segmentation model accurately segmented the caudate, putamen, and GP with high mean Dice similarity coefficients of ∼ 0.80 and significantly outperformed (p<0.01) all other considered segmentation methods. Further, an objective evaluation of the proposed method on the task of quantifying regional uptake shows that the method yielded reliable quantification with low ensemble normalized root mean square error (NRMSE) < 20% for all the considered regions. In particular, the method yielded an even lower ensemble NRMSE of ∼ 10% for the caudate and putamen.
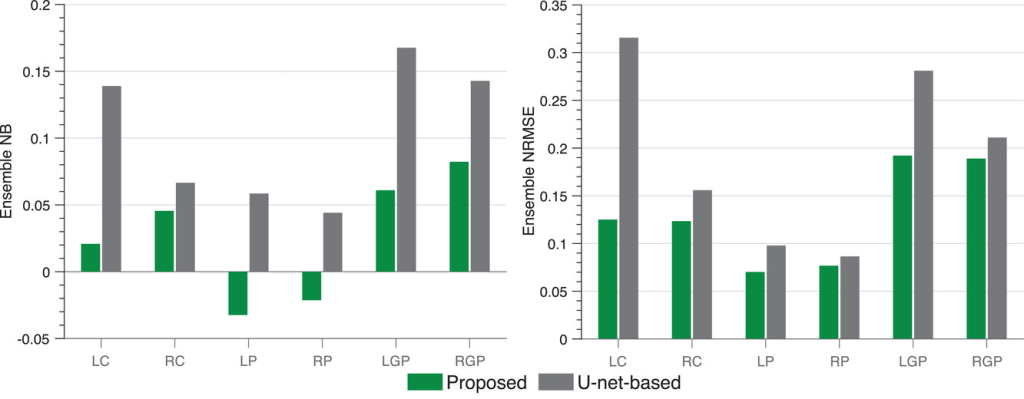
The ensemble NB and NRMSE of the estimated regional uptake yielded by the proposed method are shown in the figure below. The method reliably quantified the regional uptake with low ensemble NRMSE values (<20%) for all considered regions. In particular, the method yielded even lower ensemble NRMSE values (∼10%) for the caudate and putamen. Further, the method yielded a low ensemble NB value (<10%) for each region. Additionally, the proposed method consistently outperformed the U-net-based method on the basis of both ensemble NB and ensemble NRMSE.
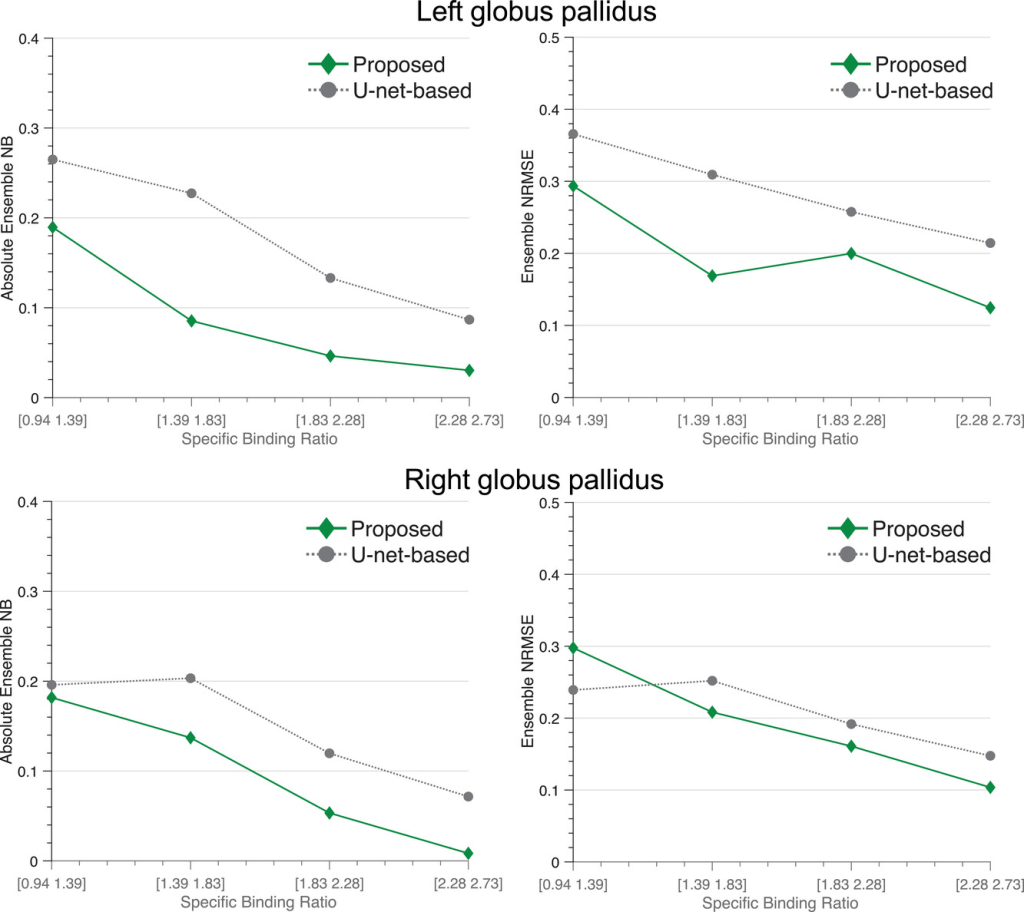
The figure below shows that for all the considered levels of SBR of the GP, the proposed method reliably quantified the uptake in the GP. On average, the proposed method yielded an ensemble NRMSE of ∼20% and an absolute ensemble NB of ∼10%. As the SBR increased, the proposed method yielded improved quantification performance with ensemble NRMSE and absolute ensemble NB approaching 10% and 0%, respectively. Additionally, the proposed method consistently yielded lower absolute ensemble NB compared to the U-net-based method.
Claim: A tissue-fraction estimation-based segmentation method was proposed for quantitative DaT SPECT. The proposed method yields fully automated segmentation of the caudate, putamen, and GP by estimating the fractional volumes occupied by these regions within each voxel of a 3D DaT-SPECT image.
Evaluations using clinically guided highly realistic simulation studies demonstrated the ability of the method to accurately segment these regions. Additionally, the proposed method significantly outperformed all other considered segmentation methods, including a state-of-the-art U-net-based method. Further, on the task of regional uptake quantification, the proposed method reliably estimated the uptake within the considered regions.
Overall, these results demonstrate the efficacy of the proposed method for accurate segmentation and reliable quantification from DaT-SPECT images. The results motivate further evaluation of the method with physical-phantom and patient studies.
