Task: Denoising of low-activity PET reconstruction of [11C]PiB and [18F]FE-PE2I images in neurodegenerative disorders
Description: The brainPETNR framework utilizes a Frequency Attention Network (FAN) model for denoising of low-activity reconstructed brain PET images. The brainPETNR model’s reference publication investigates the feasibility of using activity reduction in clinical [18F]FE-PE2I and [11C]PiB PET/CT scans, simulating low injected activity or scanning time reduction, in combination with AI-assisted denoising using brainPETNR. A single network architecture is introduced that can robustly learn denoising of brain images in which the tracer uptake is spanning from localized to delocalized spatial distributions.
The brainPETNR framework includes code for image preparation, model inference, segmentation and metrics extraction.
Code in postprocessing was used for:
- image inference
- image similarity metrics computation
- clinical metrics computation
Code in segmentation was used for:
- segmentations of ROIs (used for clinical metrics)
The code used for training is found at CAAI’s shared scripts GitHub repository > AttentionUNet().
The original inspiration of the code comes from the Frequency Attention Network (FAN) Image Denoising Model.
To generate standard-activity PET images from low-activity PET images, a three-dimensional U-Net model was employed (Ronneberger et al., 2015) with encoder, bridge, and decoder as shown in the figure below.

The encoder consisted of four identical blocks, which successively reduced the image resolution by downsampling, while increasing the number of filters to learn features at different layers. We implemented a modified version of the Frequency Attention Network (FAN) model (Mo et al., 2020) adapted to 3D single color channel image input. The input to the network was two-fold with the PET image and a noise map, which was obtained from passing the raw image through a convolutional neural network consisting of five identical layers of 64 filters. The basic block consisted of two repetitions of convolution with filter size 3 × 3 × 3, parametric rectified linear unit (PReLU) activation, followed by an average pooling layer with stride 2 to half the resolution. Each encoder block ended with a Spatial-Channel-Attention block (SCAB) designed for feature enhancement.
The first block in the encoder used 64 kernels, which was then doubled for every new block until a kernel size of 512 was reached in the bridge section. Inversely, the kernel size was halved for every new block up through the decoder. To retain the features learned through the encoder, the U-Net was constructed with skip connections that concatenated the upsampled feature map with the corresponding feature map of equal resolution from the encoding part, before being fed through the next decoding block.
The initial input to the network consisted of the 2 mm post-reconstruction smoothed low-activity PET, which is skull-stripped and cropped from 256 × 256 × 256 to 176 × 176 × 200 voxels. To fit the GPU memory during training, 8 random sub-volumes of 176 × 176 × 16 were extracted per volume and per epoch, one sub-volume serving as input to the network at a time. The target image was the 5 mm post-reconstruction smoothed standard-activity PET, equally prepared. Inference of the full volume was done by processing sub-volumes of 176 × 176 × 16 through the trained model. An intermediate 176 × 176 × 200 image was reconstructed from a dense grid of 24 sub-volumes with an overlap of 8 in the vertical direction. The inferred image was finally padded to recover the full 256 × 256 × 256-voxel denoised image.
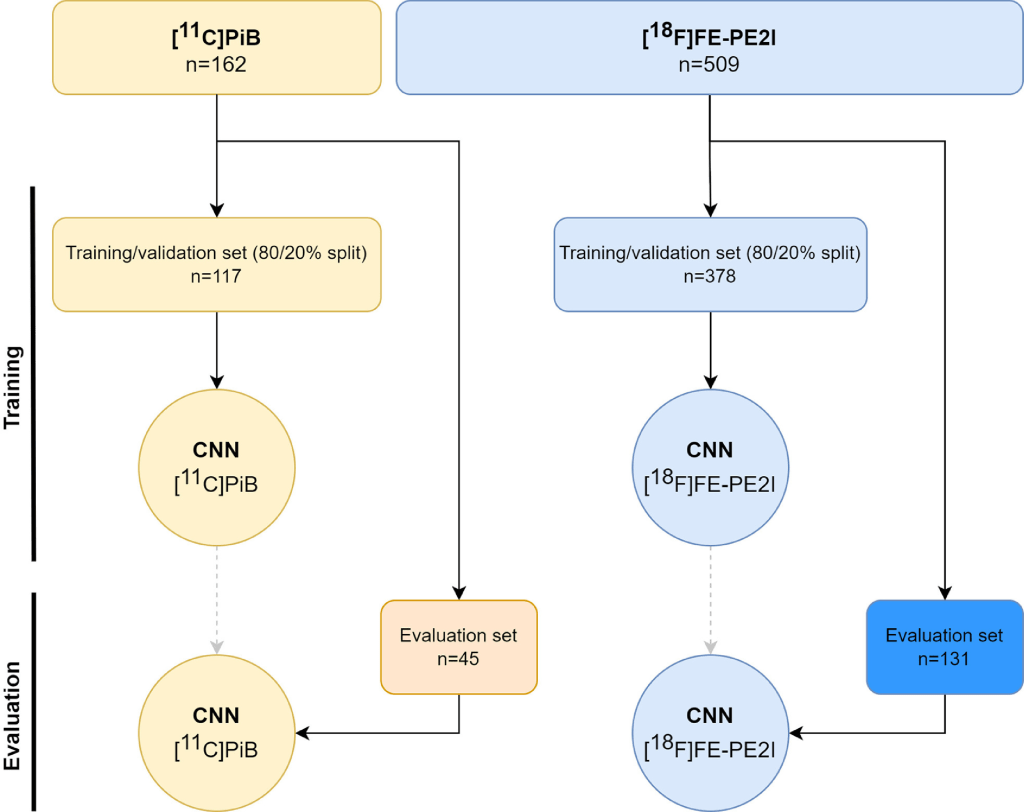
Two individual models were independently trained on their respective cohort. The data for each cohort were handled separately and split as visualized in the figure below. All following steps are done independently for each cohort. Firstly, only the newest data (post February 2021) were retained as evaluation data set to assess the performance of the two final models. All data prior to February 2021 were split into train and validation in fractions of 80% and 20%, respectively. The split in time can be considered a limited type of external validation (Kleppe et al., 2021). Each model’s precise architecture and hyperparameters were tuned using the train set with an internal validation split of 90/10%. Few of the best models for each cohort were then fed their respective validation data to derive specific clinical and noise metrics so as to find the best performing model. Finally, the best model for each cohort was used to denoise the low-activity and short-scan-time images from the evaluation data set of their respective cohort.
Data availability statement: The source code of the artificial neural network, from which the presented models are derived, preprocessing steps for model input, making inferences, and metrics calculation are available at the brainPETNR model’s reference GitHub page. The code also allows for training with transfer learning from the shared models on local data. Local data permission does not allow sharing the training and test patient data.
Results: The low-activity PET images were visually improved by the denoising model. The figure below shows examples of typical normal, borderline, and abnormal PET scans for both cohorts. The denoised images are smoothed beyond the level of the standard-activity images, which was an expected effect of the denoising algorithm (Gong et al., 2018). The over-smoothing is however minimized by using 2 mm-smoothed images as input and choosing an appropriate loss function (mean absolute error) during training.
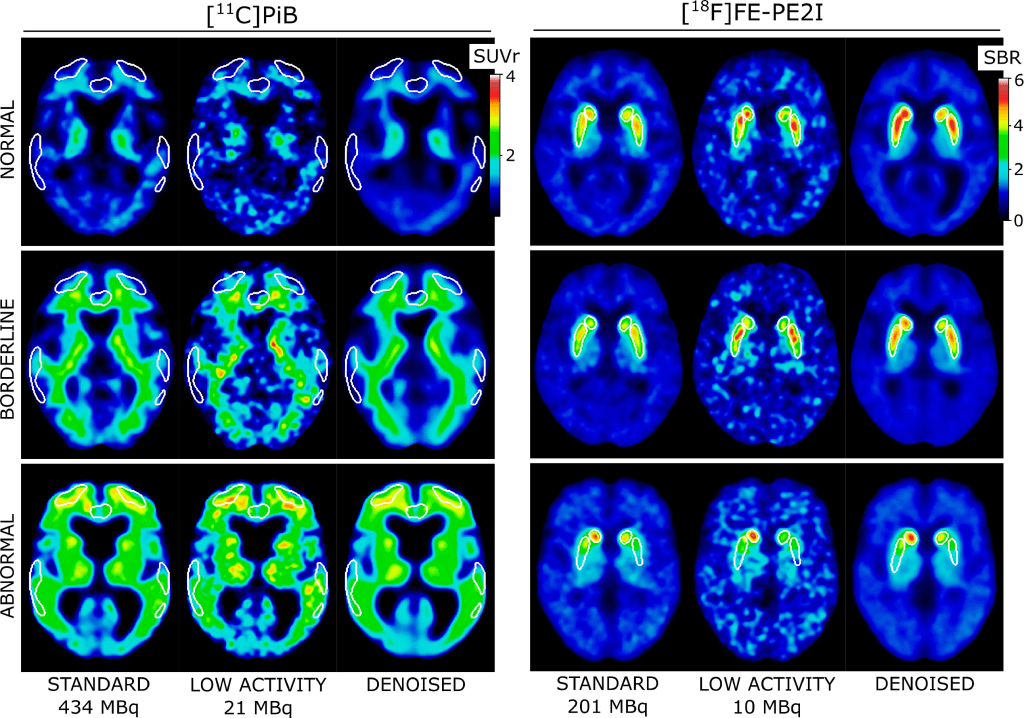
In brainPETNR model’s reference study it has been shown that [11C]PiB and [18F]FE-PE2I perform well at low activities and subsequent denoising of the acquired PET images. Image quality can be restored, while clinically relevant metrics remain within 2% of the true values. These tracers may be suitable for use in PET imaging with reduced activity or scanning time in combination with AI enhancement in the near future. Remarkably, each presented model is contained within the data acquired from a stand-alone clinical PET/CT scanner. This obviates the need for additional MRI data and issues with vendor- or sequence-specific differences. Simultaneously it minimizes data management, thereby creating a setup that is better integrated into a clinical workflow.
The presented network represents a robust architecture for denoising PET images of the brain. The two independent models trained for different clinical conditions and uptake signatures (localized and de-localized) are efficient at denoising low-activity PET images to high-quality standard images. This network is, therefore, a strong candidate for investigating reduced-activity brain PET imaging with other tracers or scanner systems by transfer learning from the supplied models.
Claim: The brainPETNR model is a deep-learning assisted denoising framework designed for low-activity [11C]PiB and [18F]FE-PE2I brain PET scans for patients with uncertain Alzheimer’s disease and Parkinson’s disease, respectively. A major activity reduction to 5% of standard activity can be achieved for both tracers without significant loss of diagnostic accuracy after processing the noisy low-activity images through the presented models. The AI models improved image quality and preserved the critical metrics used in clinical diagnosis. Future work on these cohorts should include careful clinical evaluation of low-activity, standard-activity, and denoised images, with particular care put on borderline patients. Activity reduction for PET imaging is relevant in the health care system for patient and staff protection against radiation, reducing costs, and using tracers and instrumentation more efficiently.
Frequency Attention Network (FAN) Denoising Model
Shared scripts by the CAAI research group (Rigshospitalet, University of Copenhagen, Denmark) for training deep learning networks in torch.
