Neuroimaging for the Prevention of Alzheimer’s Disease
NiftyPAD is a freely available, open source, Python-based software package for the versatile and quantitative analyses of static, full or dual-time window dynamic brain PET data.
The key novelties of NiftyPAD are the analyses of dual-time window scans with reference input processing, pharmacokinetic modelling with shortened PET acquisitions through the incorporation of arterial spin labelling (ASL)-derived relative perfusion measures, as well as optional PET data-based motion correction.
NiftyPAD is a lightweight package with minimal dependencies supporting multiple platforms with modular setup to facilitate addition of new functionalities, a user-friendly interface & integration to processing pipelines.
Features
NiftyPAD was designed to support several important features which are not available in other existing software packages for kinetic modeling:
- Freely available (Python-based) and open-source, allowing for full transparency
- Analysis of static, dynamic and dual-time window PET data
- Pharmacokinetic modelling with the incorporation of arterial spin labelling (ASL)-derived relative perfusion measures for simultaneous PET-MR scans
- Motion correction, through a built-in kinetics based realignment function, and options for excluding PET frames with large motion-misaligned attenuation correction in kinetic analysis
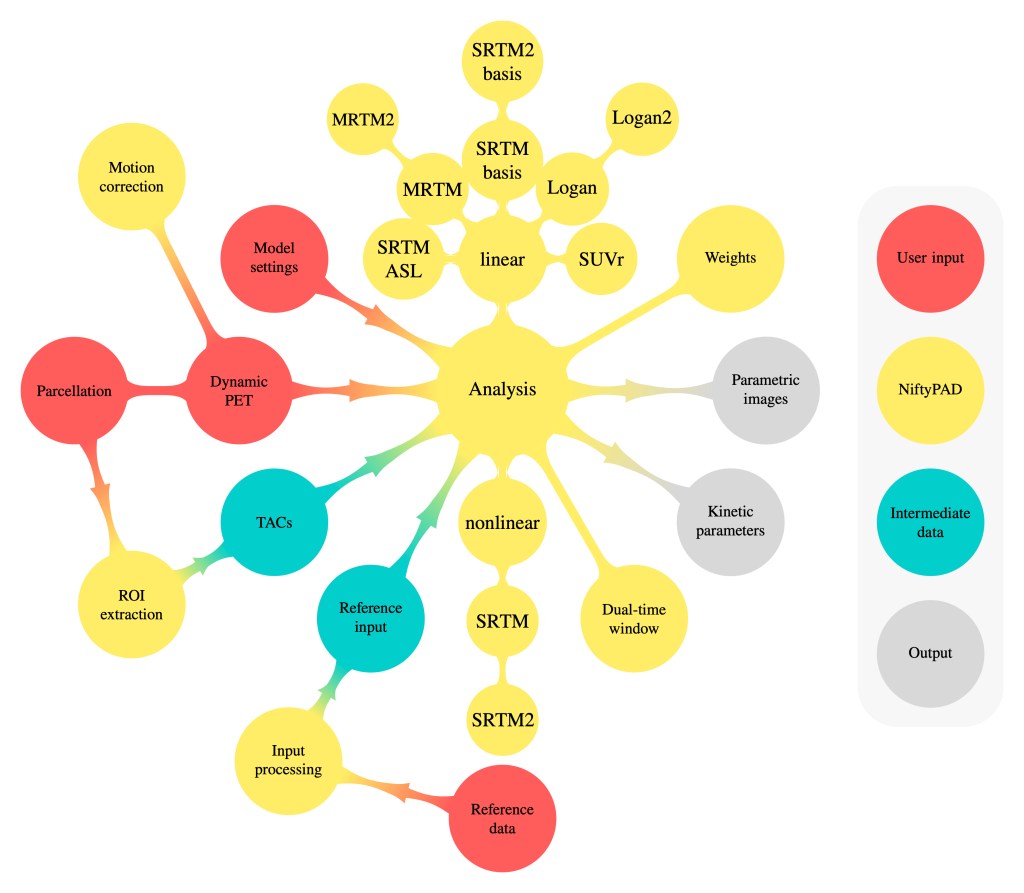
The figure below [1] summarizes the features of NiftyPAD and its workflow:
NiftyPAD provides a group of reference-based kinetic models to generate parametric images or regional kinetic parameters. For quantification, NiftyPAD requires the user to provide a PET scan, data for obtaining a reference tissue time-activity curve (TAC), a region of interest (ROI) template in case of regional analyses, and modelling settings for the selected kinetic models.
With regard to the PET scan, most commonly used imaging data formats are supported, such as dicom, nifti, analyze and ecat (see the NiBabel Python library for a full list of supported file types). In addition, fitting settings (e.g. the number of basis functions and the applied boundary conditions) can be specified.
More specifically, reference tissue input processing is implemented (i) for interpolating missing reference tissue data points in the specific case of dual-time window scans where the dynamic data are acquired in separate sessions, and (ii) to improve accuracy of the reference tissue curve in case of noise or motion. Other implemented features include support for weighting the temporal data points and options for motion correction.
Pharmacokinetic Quantification
The current version of NiftyPAD focuses on a group of reference tissue input based methods that do not require invasive arterial blood sampling or processing, which are widely applied to a range of neuro studies LaMontagne et al. (2019), Golla et al. (2016), Mertens et al. (2020).
The code for all methods is available at the GitHub page (models.py) . The kinetic parameters of interest are estimated by numerical optimization, using weighted (unweighted if weights are not given) least squares as the objective function. For quantification of both region of interest (ROI) and voxel data, the following linear and nonlinear models are implemented in NiftyPAD:
Nonlinear Models
- SRTM & SRTM2
Linear Models
- SRTM Basis & SRTM2 Basis
- SRTM ASL
- LoganRef & LoganRef2
- MRTM & MRTM2
Semi-quantitative Analysis
- Standardized uptake value ratio (SUVr) is calculated as the ratio of target to reference tissue activity for a pre-defined time window.
Weighting Schemes
- a number of weighting schemes (implemented at weights.py) are supported to facilitate optimal fitting according to Yaqub et al. (2006),
- user-defined weighting schemes are also supported
The software is open-source and distribute under the Apache-2.0 license:
Reference (citation for users/developers)
[1] Jiao, J., Heeman, F., Dixon, R. et al. NiftyPAD – Novel Python Package for Quantitative Analysis of Dynamic PET Data. Neuroinform 21, 457–468 (2023) DOI:10.1007/s12021-022-09616-0